Regulation of neural circuit formation by protocadherins
- PMID: 28631008
- PMCID: PMC5643215
- DOI: 10.1007/s00018-017-2572-3
Regulation of neural circuit formation by protocadherins
Abstract
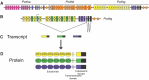
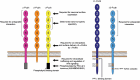
The protocadherins (Pcdhs), which make up the most diverse group within the cadherin superfamily, were first discovered in the early 1990s. Data implicating the Pcdhs, including ~60 proteins encoded by the tandem Pcdha, Pcdhb, and Pcdhg gene clusters and another ~10 non-clustered Pcdhs, in the regulation of neural development have continually accumulated, with a significant expansion of the field over the past decade. Here, we review the many roles played by clustered and non-clustered Pcdhs in multiple steps important for the formation and function of neural circuits, including dendrite arborization, axon outgrowth and targeting, synaptogenesis, and synapse elimination. We further discuss studies implicating mutation or epigenetic dysregulation of Pcdh genes in a variety of human neurodevelopmental and neurological disorders. With recent structural modeling of Pcdh proteins, the prospects for uncovering molecular mechanisms of Pcdh extracellular and intracellular interactions, and their role in normal and disrupted neural circuit formation, are bright.
Keywords: Axon outgrowth; Axonal targeting; Cell adhesion molecules; Dendrites; Dendritic arborization; Disease; Synapse elimination; Synaptogenesis.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

