Methods of probing the interactions between small molecules and disordered proteins
- PMID: 28631009
- PMCID: PMC5533867
- DOI: 10.1007/s00018-017-2563-4
Methods of probing the interactions between small molecules and disordered proteins
Abstract
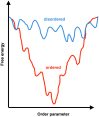
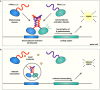
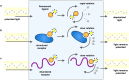
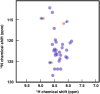
It is generally recognized that a large fraction of the human proteome is made up of proteins that remain disordered in their native states. Despite the fact that such proteins play key biological roles and are involved in many major human diseases, they still represent challenging targets for drug discovery. A major bottleneck for the identification of compounds capable of interacting with these proteins and modulating their disease-promoting behaviour is the development of effective techniques to probe such interactions. The difficulties in carrying out binding measurements have resulted in a poor understanding of the mechanisms underlying these interactions. In order to facilitate further methodological advances, here we review the most commonly used techniques to probe three types of interactions involving small molecules: (1) those that disrupt functional interactions between disordered proteins; (2) those that inhibit the aberrant aggregation of disordered proteins, and (3) those that lead to binding disordered proteins in their monomeric states. In discussing these techniques, we also point out directions for future developments.
Keywords: Binding; Disordered proteins; Drugs; Molecular interactions; Small molecules.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

