Loss of Sendai virus C protein leads to accumulation of RIG-I immunostimulatory defective interfering RNA
- PMID: 28631605
- PMCID: PMC5962894
- DOI: 10.1099/jgv.0.000815
Loss of Sendai virus C protein leads to accumulation of RIG-I immunostimulatory defective interfering RNA
Abstract
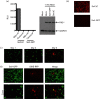
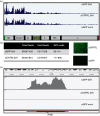
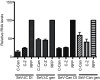
Retinoic acid inducible gene (RIG-I)-mediated innate immunity plays a pivotal role in defence against virus infections. Previously we have shown that Sendai virus (SeV) defective interfering (DI) RNA functions as an exclusive and potent RIG-I ligand in DI-RNA-rich SeV-Cantell infected cells. To further understand how RIG-I is activated during SeV infection, we used a different interferon (IFN)-inducing SeV strain, recombinant SeVΔC, which, in contrast to SeV-Cantell is believed to stimulate IFN production due to the lack of the SeV IFN antagonist protein C. Surprisingly, we found that in SevΔC-infected cells, DI RNAs also functioned as an exclusive RIG-I ligand. Infections with wild-type SeV failed to generate any RIG-I-associated immunostimulatory RNA and this correlated with the lack of DI genomes in infected cells, as well as with the absence of cellular innate immune responses. Supplementation of the C protein in the context of SeVΔC infection led to a reduction in the number of DI RNAs, further supporting the potential role of the C protein as a negative regulator of DI generation and/or accumulation. Our findings indicate that limiting DI genome production is an important function of viral IFN antagonist proteins.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

