A Comparative Study of the Structural Dynamics of Four Terminal Uridylyl Transferases
- PMID: 28632168
- PMCID: PMC5485530
- DOI: 10.3390/genes8060166
A Comparative Study of the Structural Dynamics of Four Terminal Uridylyl Transferases
Abstract
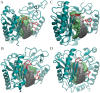
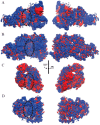
African trypanosomiasis occurs in 36 countries in sub-Saharan Africa with 10,000 reported cases annually. No definitive remedy is currently available and if left untreated, the disease becomes fatal. Structural and biochemical studies of trypanosomal terminal uridylyl transferases (TUTases) demonstrated their functional role in extensive uridylate insertion/deletion of RNA. Trypanosoma brucei RNA Editing TUTase 1 (TbRET1) is involved in guide RNA 3' end uridylation and maturation, while TbRET2 is responsible for U-insertion at RNA editing sites. Two additional TUTases called TbMEAT1 and TbTUT4 have also been reported to share similar function. TbRET1 and TbRET2 are essential enzymes for the parasite viability making them potential drug targets. For this study, we clustered molecular dynamics (MD) trajectories of four TUTases based on active site shape measured by Pocket Volume Measurer (POVME) program. Among the four TUTases, TbRET1 exhibited the largest average pocket volume, while TbMEAT1's and TbTUT4's active sites displayed the most flexibility. A side pocket was also identified within the active site in all TUTases with TbRET1 having the most pronounced. Our results indicate that TbRET1's larger side pocket can be exploited to achieve selective inhibitor design as FTMap identifies it as a druggable pocket.
Keywords: MEAT1; POVME; RET1; RET2; TUT4; TUTases; Trypanosoma brucei; electrostatics; pocket shape; pocket volume; terminal uridylyl transferases.
Conflict of interest statement
Rommie E. Amaro is a co-founder of, on the scientific advisory board of, and has equity interest in Actavalon, Inc.
Figures
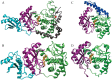

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

