Metabolomics as a Driver in Advancing Precision Medicine in Sepsis
- PMID: 28632924
- PMCID: PMC5600684
- DOI: 10.1002/phar.1974
Metabolomics as a Driver in Advancing Precision Medicine in Sepsis
Abstract
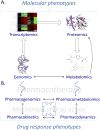
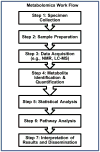
The objective of this review is to explain the science of metabolomics-a science of systems biology that measures and studies endogenous small molecules (metabolites) that are present in a single biological sample-and its application to the diagnosis and treatment of sepsis. In addition, we discuss how discovery through metabolomics can contribute to the development of precision medicine targets for this complex disease state and the potential avenues for those new discoveries to be applied in the clinical environment. A nonsystematic literature review was performed focusing on metabolomics, pharmacometabolomics, and sepsis. Human (adult and pediatric) and animal studies were included. Metabolomics has been investigated in the diagnosis, prognosis, and risk stratification of sepsis, as well as for the identification of drug target opportunities. Metabolomics elucidates a new level of detail when compared with other systems biology sciences, with regard to the metabolites that are most relevant in the pathophysiology of sepsis, as well as highlighting specific biochemical pathways at work in sepsis. Metabolomics also highlights biochemical differences between sepsis survivors and nonsurvivors at a level of detail greater than that demonstrated by genomics, transcriptomics, or proteomics, potentially leading to actionable targets for new therapies. The application of pharmacometabolomics and its integration with other systems pharmacology to sepsis therapeutics could be particularly helpful in differentiating drug responders and nonresponders and furthering knowledge of mechanisms of drug action and response. The accumulated literature on metabolomics suggests it is a viable tool for continued discovery around the pathophysiology, diagnosis and prognosis, and treatment of sepsis in both adults and children, and it provides a greater level of biochemical detail and insight than other systems biology approaches. However, the clinical application of metabolomics in sepsis has not yet been fully realized. Prospective validation studies are needed to translate metabolites from the discovery phase into the clinical utility phase.
Keywords: critical care; pediatrics; pharmacometabolomics; pharmacotherapy; systems biology; systems pharmacology.
© 2017 Pharmacotherapy Publications, Inc.
Figures
References
-
- Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med. 2016;3:259–72. - PubMed
-
- Vincent JL, Marshall JC, Namendys-Silva SA, et al. Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir Med. 2014;5:380–6. - PubMed
-
- Watson RS, Carcillo JA. Scope and epidemiology of pediatric sepsis. Pediatr Crit Care Med. 2005;3(1):S3–5. - PubMed
-
- Bryce J, Boschi-Pinto C, Shibuya K, Black RE Group WCHER. WHO estimates of the causes of death in children. Lancet. 2005;9465:1147–52. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

