Structured Populations of Sulfolobus acidocaldarius with Susceptibility to Mobile Genetic Elements
- PMID: 28633403
- PMCID: PMC5554439
- DOI: 10.1093/gbe/evx104
Structured Populations of Sulfolobus acidocaldarius with Susceptibility to Mobile Genetic Elements
Abstract
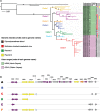
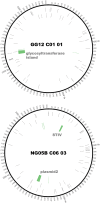
The impact of a structured environment on genome evolution can be determined through comparative population genomics of species that live in the same habitat. Recent work comparing three genome sequences of Sulfolobus acidocaldarius suggested that highly structured, extreme, hot spring environments do not limit dispersal of this thermoacidophile, in contrast to other co-occurring Sulfolobus species. Instead, a high level of conservation among these three S. acidocaldarius genomes was hypothesized to result from rapid, global-scale dispersal promoted by low susceptibility to viruses that sets S. acidocaldarius apart from its sister Sulfolobus species. To test this hypothesis, we conducted a comparative analysis of 47 genomes of S. acidocaldarius from spatial and temporal sampling of two hot springs in Yellowstone National Park. While we confirm the low diversity in the core genome, we observe differentiation among S. acidocaldarius populations, likely resulting from low migration among hot spring "islands" in Yellowstone National Park. Patterns of genomic variation indicate that differing geological contexts result in the elimination or preservation of diversity among differentiated populations. We observe multiple deletions associated with a large genomic island rich in glycosyltransferases, differential integrations of the Sulfolobus turreted icosahedral virus, as well as two different plasmid elements. These data demonstrate that neither rapid dispersal nor lack of mobile genetic elements result in low diversity in the S. acidocaldarius genomes. We suggest instead that significant differences in the recent evolutionary history, or the intrinsic evolutionary rates, of sister Sulfolobus species result in the relatively low diversity of the S. acidocaldarius genome.
Keywords: Sulfolobus species; genome divergence; geothermal habitat; immigration; pangenome; population dynamics.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

