Genetic Indicators of Drug Resistance in the Highly Repetitive Genome of Trichomonas vaginalis
- PMID: 28633446
- PMCID: PMC5522705
- DOI: 10.1093/gbe/evx110
Genetic Indicators of Drug Resistance in the Highly Repetitive Genome of Trichomonas vaginalis
Abstract
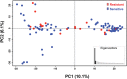
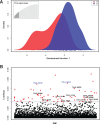
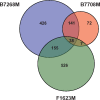
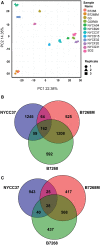
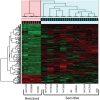
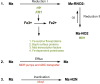
Trichomonas vaginalis, the most common nonviral sexually transmitted parasite, causes ∼283 million trichomoniasis infections annually and is associated with pregnancy complications and increased risk of HIV-1 acquisition. The antimicrobial drug metronidazole is used for treatment, but in a fraction of clinical cases, the parasites can become resistant to this drug. We undertook sequencing of multiple clinical isolates and lab derived lines to identify genetic markers and mechanisms of metronidazole resistance. Reduced representation genome sequencing of ∼100 T. vaginalis clinical isolates identified 3,923 SNP markers and presence of a bipartite population structure. Linkage disequilibrium was found to decay rapidly, suggesting genome-wide recombination and the feasibility of genetic association studies in the parasite. We identified 72 SNPs associated with metronidazole resistance, and a comparison of SNPs within several lab-derived resistant lines revealed an overlap with the clinically resistant isolates. We identified SNPs in genes for which no function has yet been assigned, as well as in functionally-characterized genes relevant to drug resistance (e.g., pyruvate:ferredoxin oxidoreductase). Transcription profiles of resistant strains showed common changes in genes involved in drug activation (e.g., flavin reductase), accumulation (e.g., multidrug resistance pump), and detoxification (e.g., nitroreductase). Finally, we identified convergent genetic changes in lab-derived resistant lines of Tritrichomonas foetus, a distantly related species that causes venereal disease in cattle. Shared genetic changes within and between T. vaginalis and Tr. foetus parasites suggest conservation of the pathways through which adaptation has occurred. These findings extend our knowledge of drug resistance in the parasite, providing a panel of markers that can be used as a diagnostic tool.
Keywords: Trichomonas vaginalis; antimicrobial drug resistance; comparative genomics; genetic association study; sexually transmitted infection.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Barratt J, Gough R, Stark D, Ellis J.. 2016. Bulky trichomonad genomes: encoding a Swiss Army knife. Trends Parasitol. 32:783–797. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

