Epigenetic silencing of downstream genes mediated by tandem orientation in lung cancer
- PMID: 28634396
- PMCID: PMC5478622
- DOI: 10.1038/s41598-017-04248-w
Epigenetic silencing of downstream genes mediated by tandem orientation in lung cancer
Abstract
Epigenetic deregulation is of importance in tumorigenesis. In particular CpG islands (CGI), are frequently hypermethylated. Here, genome-wide DNA-methylation profiles of 480,000 CpGs in lung cancer cells were generated. It was observed that intra- and intergenic CGI exhibited higher methylation compared to normal cells. The functional annotation of hypermethylated CGI revealed that the hypermethylation was associated with homeobox domain genes and targets marked by repressive histone modifications. The strongest methylation variation was observed in transitional areas of CGI, termed shores. 5'-shores of promoter-associated CGI in lung cancer cell lines were higher methylated than 3'-shores. Within two tandem-oriented genes, a significant hypermethylation of the downstream-located CGI promoters was revealed. Hypermethylation correlates with the length of the intergenic region between such tandem genes. As the RASSF1A tumor suppressor gene represents such a downstream tandem gene, its silencing was analyzed using an inducible system. It was determined that the induction of an upstream gene led to a repression of RASSF1A through a process involving histone deacetylases and CPSF1. A tumor-specific increase in expression of histone deacetylases and CPSF1 was detected in lung cancer. Our results suggest that the downstream gene could be susceptible to epigenetic silencing when organized in a tandem orientation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


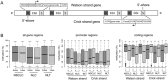

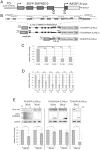
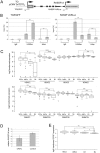
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

