Regulation of Escherichia coli Pathogenesis by Alternative Sigma Factor N
- PMID: 28635589
- PMCID: PMC11575691
- DOI: 10.1128/ecosalplus.ESP-0016-2016
Regulation of Escherichia coli Pathogenesis by Alternative Sigma Factor N
Abstract
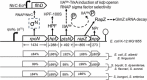
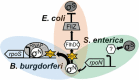
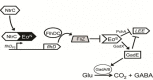
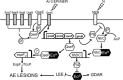
σN (also σ54) is an alternative sigma factor subunit of the RNA polymerase complex that regulates the expression of genes from many different ontological groups. It is broadly conserved in the Eubacteria with major roles in nitrogen metabolism, membrane biogenesis, and motility. σN is encoded as the first gene of a five-gene operon including rpoN (σN), ptsN, hpf, rapZ, and npr that has been genetically retained among species of Escherichia, Shigella, and Salmonella. In an increasing number of bacteria, σN has been implicated in the control of genes essential to pathogenic behavior, including those involved in adherence, secretion, immune subversion, biofilm formation, toxin production, and resistance to both antimicrobials and biological stressors. For most pathogens how this is achieved is unknown. In enterohemorrhagic Escherichia coli (EHEC) O157, Salmonella enterica, and Borrelia burgdorferi, regulation of virulence by σN requires another alternative sigma factor, σS, yet the model by which σN-σS virulence regulation is predicted to occur is varied in each of these pathogens. In this review, the importance of σN to bacterial pathogenesis is introduced, and common features of σN-dependent virulence regulation discussed. Emphasis is placed on the molecular mechanisms underlying σN virulence regulation in E. coli O157. This includes a review of the structure and function of regulatory pathways connecting σN to virulence expression, predicted input signals for pathway stimulation, and the role for cognate σN activators in initiation of gene systems determining pathogenic behavior.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

