A Review of Flaviviruses that Have No Known Arthropod Vector
- PMID: 28635667
- PMCID: PMC5490829
- DOI: 10.3390/v9060154
A Review of Flaviviruses that Have No Known Arthropod Vector
Abstract
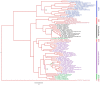
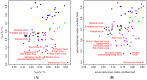
Most viruses in the genus Flavivirus are horizontally transmitted between hematophagous arthropods and vertebrate hosts, but some are maintained in arthropod- or vertebrate-restricted transmission cycles. Flaviviruses maintained by vertebrate-only transmission are commonly referred to as no known vector (NKV) flaviviruses. Fourteen species and two subtypes of NKV flaviviruses are recognized by the International Committee on Taxonomy of Viruses (ICTV), and Tamana bat virus potentially belongs to this group. NKV flaviviruses have been isolated in nature almost exclusively from bats and rodents; exceptions are the two isolates of Dakar bat virus recovered from febrile humans and the recent isolations of Sokoluk virus from field-collected ticks, which raises questions as to whether it should remain classified as an NKV flavivirus. There is evidence to suggest that two other NKV flaviviruses, Entebbe bat virus and Yokose virus, may also infect arthropods in nature. The best characterized bat- and rodent-associated NKV flaviviruses are Rio Bravo and Modoc viruses, respectively, but both have received limited research attention compared to many of their arthropod-infecting counterparts. Herein, we provide a comprehensive review of NKV flaviviruses, placing a particular emphasis on their classification, host range, geographic distribution, replication kinetics, pathogenesis, transmissibility and molecular biology.
Keywords: bat; flavivirus; genomic organization; host range; no known vector; rodent; transmission; vertebrate-specific.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lindenbach B.D., Murray C.L., Thiel H.-J., Rice C.M. Flaviviridae. In: Knipe D.M., Howley P.M., editors. Fields Virolgy. Lippincott Williams and Wilkins; Philadelphia, PA, USA: 2013. pp. 712–746.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

