Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis
- PMID: 28635960
- PMCID: PMC5482061
- DOI: 10.1038/ncomms15869
Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis
Abstract
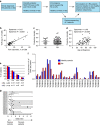
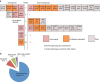
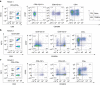
Somatic mutations contribute to tumorigenesis. Although these mutations occur in all proliferating cells, their accumulation under non-malignant conditions, such as in autoimmune disorders, has not been investigated. Here, we show that patients with newly diagnosed rheumatoid arthritis have expanded CD8+ T-cell clones; in 20% (5/25) of patients CD8+ T cells, but not CD4+ T cells, harbour somatic mutations. In healthy controls (n=20), only one mutation is identified in the CD8+ T-cell pool. Mutations exist exclusively in the expanded CD8+ effector-memory subset, persist during follow-up, and are predicted to change protein functions. Some of the mutated genes (SLAMF6, IRF1) have previously been associated with autoimmunity. RNA sequencing of mutation-harbouring cells shows signatures corresponding to cell proliferation. Our data provide evidence of accumulation of somatic mutations in expanded CD8+ T cells, which may have pathogenic significance for RA and other autoimmune diseases.
Conflict of interest statement
S.M. and K.P. have received honoraria and research funding from Novartis, Bristol-Myers Squibb and Pfizer. S.M. has received research funding from Ariad. J.S. has received lecture fees from Roche. The remaining authors declare no competing financial interests.
Figures



References
-
- Araten D. J. et al. A quantitative measurement of the human somatic mutation rate. Cancer Res. 65, 8111–8117 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

