The metagenomic data life-cycle: standards and best practices
- PMID: 28637310
- PMCID: PMC5737865
- DOI: 10.1093/gigascience/gix047
The metagenomic data life-cycle: standards and best practices
Abstract
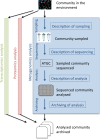
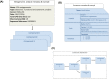
Metagenomics data analyses from independent studies can only be compared if the analysis workflows are described in a harmonized way. In this overview, we have mapped the landscape of data standards available for the description of essential steps in metagenomics: (i) material sampling, (ii) material sequencing, (iii) data analysis, and (iv) data archiving and publishing. Taking examples from marine research, we summarize essential variables used to describe material sampling processes and sequencing procedures in a metagenomics experiment. These aspects of metagenomics dataset generation have been to some extent addressed by the scientific community, but greater awareness and adoption is still needed. We emphasize the lack of standards relating to reporting how metagenomics datasets are analysed and how the metagenomics data analysis outputs should be archived and published. We propose best practice as a foundation for a community standard to enable reproducibility and better sharing of metagenomics datasets, leading ultimately to greater metagenomics data reuse and repurposing.
Keywords: best practice; data analysis; metadata; metagenomics; sampling; sequencing; standard.
© The Author 2017. Published by Oxford University Press.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

