Outbreak tracking of Aleutian mink disease virus (AMDV) using partial NS1 gene sequencing
- PMID: 28637462
- PMCID: PMC5480136
- DOI: 10.1186/s12985-017-0786-5
Outbreak tracking of Aleutian mink disease virus (AMDV) using partial NS1 gene sequencing
Abstract
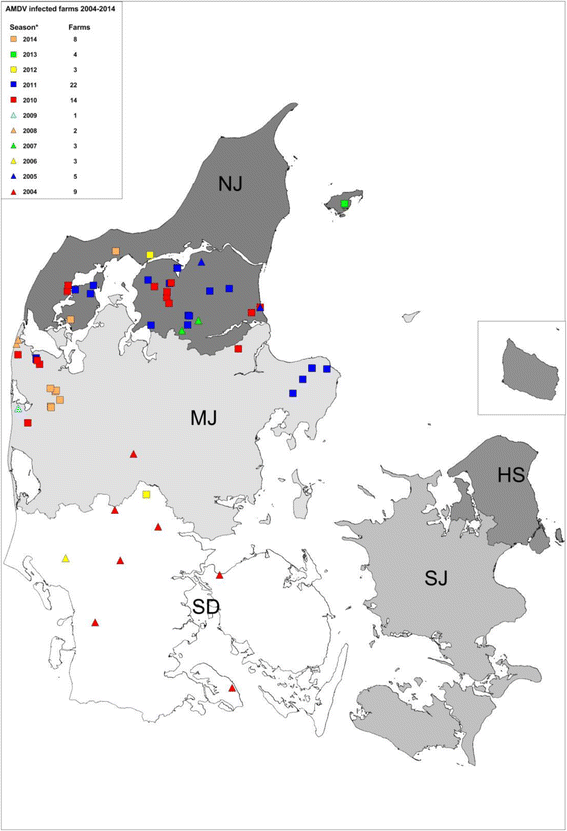
Background: Aleutian Mink Disease (AMD) is an infectious disease of mink (Neovison vison) and globally a major cause of economic losses in mink farming. The disease is caused by Aleutian Mink Disease Virus (AMDV) that belongs to the genus Amdoparvovirus within the Parvoviridae family. Several strains have been described with varying virulence and the severity of infection also depends on the host's genotype and immune status. Clinical signs include respiratory distress in kits and unthriftiness and low quality of the pelts. The infection can also be subclinical. Systematic control of AMDV in Danish mink farms was voluntarily initiated in 1976. Over recent decades the disease was mainly restricted to the very northern part of the country (Northern Jutland), with only sporadic outbreaks outside this region. Most of the viruses from this region have remained very closely related at the nucleotide level for decades. However, in 2015, several outbreaks of AMDV occurred at mink farms throughout Denmark, and the sources of these outbreaks were not known.
Methods: Partial NS1 gene sequencing, phylogenetic analyses data were utilized along with epidemiological to determine the origin of the outbreaks.
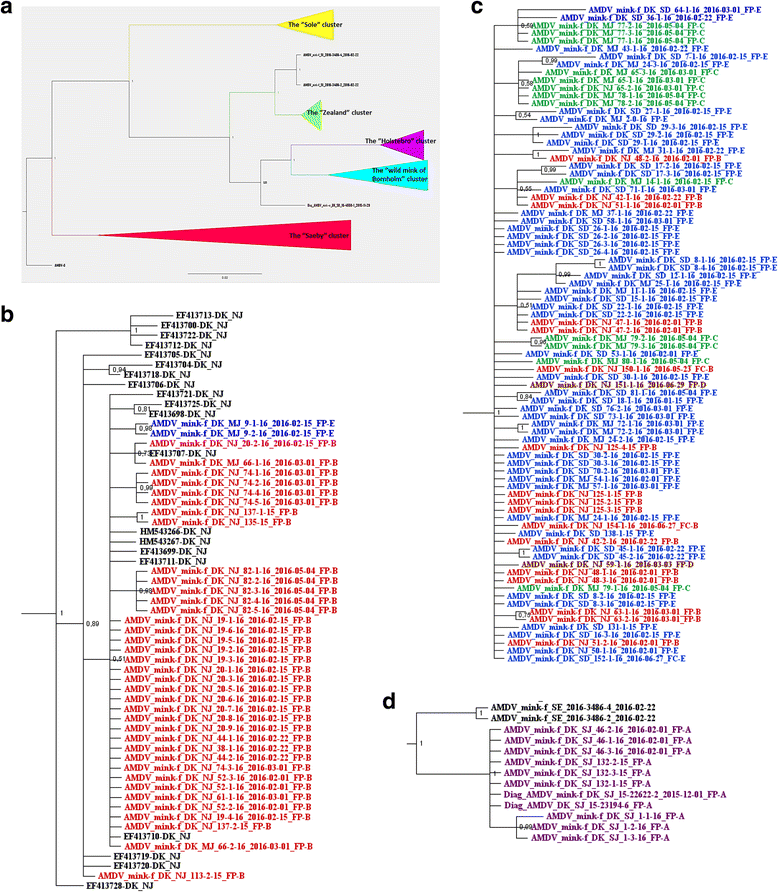
Results: The phylogenetic analyses of partial NS1 gene sequences revealed that the outbreaks were caused by two different clusters of viruses that were clearly different from the strains found in Northern Jutland. These clusters had restricted geographical distribution, and the variation within the clusters was remarkably low. The outbreaks on Zealand were epidemiologically linked and a close sequence match was found to two virus sequences from Sweden. The other cluster of outbreaks restricted to Jutland and Funen were linked to three feed producers (FP) but secondary transmissions between farms in the same geographical area could not be excluded.
Conclusion: This study confirmed that partial NS1 sequencing can be used in outbreak tracking to determine major viral clusters of AMDV. Using this method, two new distinct AMDV clusters with low intra-cluster sequence diversity were identified, and epidemiological data helped to reveal possible ways of viral introduction into the affected herds.
Keywords: Aleutian mink disease; Amdv; Carnivore amdoparvovirus; Denmark; NS1; Phylogenetic analysis; Plasmacytosis.
Figures
Similar articles
-
Global phylogenetic analysis of contemporary aleutian mink disease viruses (AMDVs).Virol J. 2017 Nov 22;14(1):231. doi: 10.1186/s12985-017-0898-y. Virol J. 2017. PMID: 29166950 Free PMC article.
-
Epidemiology, pathogenesis, and diagnosis of Aleutian disease caused by Aleutian mink disease virus: A literature review with a perspective of genomic breeding for disease control in American mink (Neogale vison).Virus Res. 2023 Oct 15;336:199208. doi: 10.1016/j.virusres.2023.199208. Epub 2023 Aug 28. Virus Res. 2023. PMID: 37633597 Free PMC article. Review.
-
Diversity and stability of Aleutian mink disease virus during bottleneck transitions resulting from eradication in domestic mink in Denmark.Vet Microbiol. 2011 Apr 21;149(1-2):64-71. doi: 10.1016/j.vetmic.2010.10.016. Epub 2010 Nov 4. Vet Microbiol. 2011. PMID: 21112164
-
Evolutionary analysis of whole-genome sequences confirms inter-farm transmission of Aleutian mink disease virus.J Gen Virol. 2017 Jun;98(6):1360-1371. doi: 10.1099/jgv.0.000777. Epub 2017 Jun 13. J Gen Virol. 2017. PMID: 28612703
-
Mink Aleutian disease seroprevalence in China during 1981-2017: A systematic review and meta-analysis.Microb Pathog. 2020 Feb;139:103908. doi: 10.1016/j.micpath.2019.103908. Epub 2019 Dec 10. Microb Pathog. 2020. PMID: 31830583
Cited by
-
Asymptomatic viral infection is associated with lower host reproductive output in wild mink populations.Sci Rep. 2023 Jun 9;13(1):9390. doi: 10.1038/s41598-023-36581-8. Sci Rep. 2023. PMID: 37296209 Free PMC article.
-
Identification of a Potential mRNA-based Vaccine Candidate against the SARS-CoV-2 Spike Glycoprotein: A Reverse Vaccinology Approach.ChemistrySelect. 2022 Feb 18;7(7):e202103903. doi: 10.1002/slct.202103903. Epub 2022 Feb 17. ChemistrySelect. 2022. PMID: 35601809 Free PMC article.
-
Global phylogenetic analysis of contemporary aleutian mink disease viruses (AMDVs).Virol J. 2017 Nov 22;14(1):231. doi: 10.1186/s12985-017-0898-y. Virol J. 2017. PMID: 29166950 Free PMC article.
-
Strong selection signatures for Aleutian disease tolerance acting on novel candidate genes linked to immune and cellular responses in American mink (Neogale vison).Sci Rep. 2024 Jan 10;14(1):1035. doi: 10.1038/s41598-023-51039-7. Sci Rep. 2024. PMID: 38200094 Free PMC article.
-
Epidemiology, pathogenesis, and diagnosis of Aleutian disease caused by Aleutian mink disease virus: A literature review with a perspective of genomic breeding for disease control in American mink (Neogale vison).Virus Res. 2023 Oct 15;336:199208. doi: 10.1016/j.virusres.2023.199208. Epub 2023 Aug 28. Virus Res. 2023. PMID: 37633597 Free PMC article. Review.
References
-
- Bloom M. Aleutian mink disease - puzzles and paradignms. Infect Agents Dis Issues Comment. 1994;3:279–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

