Niche-mediated depletion of the normal hematopoietic stem cell reservoir by Flt3-ITD-induced myeloproliferation
- PMID: 28637883
- PMCID: PMC5502426
- DOI: 10.1084/jem.20161418
Niche-mediated depletion of the normal hematopoietic stem cell reservoir by Flt3-ITD-induced myeloproliferation
Abstract
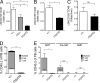
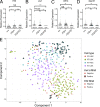
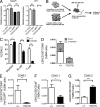
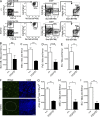
Although previous studies suggested that the expression of FMS-like tyrosine kinase 3 (Flt3) initiates downstream of mouse hematopoietic stem cells (HSCs), FLT3 internal tandem duplications (FLT3 ITDs) have recently been suggested to intrinsically suppress HSCs. Herein, single-cell interrogation found Flt3 mRNA expression to be absent in the large majority of phenotypic HSCs, with a strong negative correlation between Flt3 and HSC-associated gene expression. Flt3-ITD knock-in mice showed reduced numbers of phenotypic HSCs, with an even more severe loss of long-term repopulating HSCs, likely reflecting the presence of non-HSCs within the phenotypic HSC compartment. Competitive transplantation experiments established that Flt3-ITD compromises HSCs through an extrinsically mediated mechanism of disrupting HSC-supporting bone marrow stromal cells, with reduced numbers of endothelial and mesenchymal stromal cells showing increased inflammation-associated gene expression. Tumor necrosis factor (TNF), a cell-extrinsic potent negative regulator of HSCs, was overexpressed in bone marrow niche cells from FLT3-ITD mice, and anti-TNF treatment partially rescued the HSC phenotype. These findings, which establish that Flt3-ITD-driven myeloproliferation results in cell-extrinsic suppression of the normal HSC reservoir, are of relevance for several aspects of acute myeloid leukemia biology.
© 2017 Mead et al.
Figures


References
-
- Adolfsson J., Borge O.J., Bryder D., Theilgaard-Mönch K., Astrand-Grundström I., Sitnicka E., Sasaki Y., and Jacobsen S.E.. 2001. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 15:659–669. 10.1016/S1074-7613(01)00220-5 - DOI - PubMed
-
- Adolfsson J., Månsson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C.T., Bryder D., Yang L., Borge O.J., Thoren L.A., et al. 2005. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 121:295–306. 10.1016/j.cell.2005.02.013 - DOI - PubMed
-
- Böiers C., Buza-Vidas N., Jensen C.T., Pronk C.J., Kharazi S., Wittmann L., Sitnicka E., Hultquist A., and Jacobsen S.E.. 2010. Expression and role of FLT3 in regulation of the earliest stage of normal granulocyte-monocyte progenitor development. Blood. 115:5061–5068. 10.1182/blood-2009-12-258756 - DOI - PubMed
-
- Böiers C., Carrelha J., Lutteropp M., Luc S., Green J.C., Azzoni E., Woll P.S., Mead A.J., Hultquist A., Swiers G., et al. 2013. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 13:535–548. 10.1016/j.stem.2013.08.012 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- MR/M00919X/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_12020/MRC_/Medical Research Council/United Kingdom
- G0902418/MRC_/Medical Research Council/United Kingdom
- MC_UU_00016/5/MRC_/Medical Research Council/United Kingdom
- G0701761/MRC_/Medical Research Council/United Kingdom
- MR/L006340/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/5/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_UU_00016/7/MRC_/Medical Research Council/United Kingdom
- G0900892/MRC_/Medical Research Council/United Kingdom
- 11-0724/AICR_/Worldwide Cancer Research/United Kingdom
- G0801073/MRC_/Medical Research Council/United Kingdom
- G0501838/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/7/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

