A Novel Bromodomain Inhibitor Reverses HIV-1 Latency through Specific Binding with BRD4 to Promote Tat and P-TEFb Association
- PMID: 28638377
- PMCID: PMC5461361
- DOI: 10.3389/fmicb.2017.01035
A Novel Bromodomain Inhibitor Reverses HIV-1 Latency through Specific Binding with BRD4 to Promote Tat and P-TEFb Association
Abstract
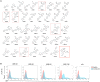
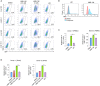
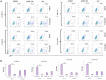
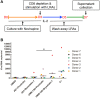
While combinatory antiretroviral therapy (cART) can effectively reduce HIV-1 viremia, it cannot eliminate HIV-1 infection. In the presence of cART, viral reservoirs remain latent, impeding the cure of HIV-1/AIDS. Recently, latency-reversing agents (LRAs) have been developed with the intent of purging latent HIV-1, providing an intriguing strategy for the eradication of the residual viral reservoirs. Our earlier studies show that the first-generation, methyl-triazolo bromodomain, and extra-terminal domain inhibitor (BETi), JQ1, facilitates the reversal of HIV-1 latency. BETis have emerged as a new class of compounds that are promising for this HIV-1 "shock and kill" eradication approach. However, when used as a single drug, JQ1 only modestly reverses HIV-1 latency, which complicates studying the underlining mechanisms. Meanwhile, it has been widely discussed that the induction of latent proviruses is stochastic (Ho et al., 2013). Thus, new BETis are currently under active development with focus on improving potency, ease of synthesis and structural diversity. Using fluorous-tagged multicomponent reactions, we developed a novel second-generation, 3,5-dimethylisoxazole BETi based on an imidazo[1,2-a] pyrazine scaffold, UMB-32. Furthermore, we screened 37 UMB-32 derivatives and identified that one, UMB-136, reactivates HIV-1 in multiple cell models of HIV-1 latency with better efficiency than either JQ1 or UMB-32. UMB-136 enhances HIV-1 transcription and increases viral production through the release of P-TEFb. Importantly, UMB-136 enhances the latency-reversing effects of PKC agonists (prostratin, bryostatin-1) in CD8-depleted PBMCs containing latent viral reservoirs. Our results illustrate that structurally improved BETis, such as UMB-136, may be useful as promising LRAs for HIV-1 eradication.
Keywords: BETi; BRD4; HIV-1; JQ1; LRA; UMB-136; latency; reactivation.
Figures

Similar articles
-
An In-Depth Comparison of Latency-Reversing Agent Combinations in Various In Vitro and Ex Vivo HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression.PLoS Pathog. 2015 Jul 30;11(7):e1005063. doi: 10.1371/journal.ppat.1005063. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26225566 Free PMC article.
-
A New Quinoline BRD4 Inhibitor Targets a Distinct Latent HIV-1 Reservoir for Reactivation from Other "Shock" Drugs.J Virol. 2018 Apr 27;92(10):e02056-17. doi: 10.1128/JVI.02056-17. Print 2018 May 15. J Virol. 2018. PMID: 29343578 Free PMC article.
-
CPI-637 as a Potential Bifunctional Latency-Reversing Agent That Targets Both the BRD4 and TIP60 Proteins.Front Cell Infect Microbiol. 2021 Jul 19;11:686035. doi: 10.3389/fcimb.2021.686035. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34350133 Free PMC article.
-
Are BET Inhibitors yet Promising Latency-Reversing Agents for HIV-1 Reactivation in AIDS Therapy?Viruses. 2021 May 29;13(6):1026. doi: 10.3390/v13061026. Viruses. 2021. PMID: 34072421 Free PMC article. Review.
-
Current Status of Latency Reversing Agents Facing the Heterogeneity of HIV-1 Cellular and Tissue Reservoirs.Front Microbiol. 2020 Jan 24;10:3060. doi: 10.3389/fmicb.2019.03060. eCollection 2019. Front Microbiol. 2020. PMID: 32038533 Free PMC article. Review.
Cited by
-
Diversity of small molecule HIV-1 latency reversing agents identified in low- and high-throughput small molecule screens.Med Res Rev. 2020 May;40(3):881-908. doi: 10.1002/med.21638. Epub 2019 Oct 13. Med Res Rev. 2020. PMID: 31608481 Free PMC article. Review.
-
Modulation of BRD4 in HIV epigenetic regulation: implications for finding an HIV cure.Retrovirology. 2021 Jan 7;18(1):3. doi: 10.1186/s12977-020-00547-9. Retrovirology. 2021. PMID: 33413475 Free PMC article. Review.
-
KDM5A/B contribute to HIV-1 latent infection and survival of HIV-1 infected cells.Antiviral Res. 2024 Aug;228:105947. doi: 10.1016/j.antiviral.2024.105947. Epub 2024 Jun 24. Antiviral Res. 2024. PMID: 38925368 Free PMC article.
-
Insights Into Persistent HIV-1 Infection and Functional Cure: Novel Capabilities and Strategies.Front Microbiol. 2022 Apr 27;13:862270. doi: 10.3389/fmicb.2022.862270. eCollection 2022. Front Microbiol. 2022. PMID: 35572626 Free PMC article. Review.
-
Bromodomains in Human-Immunodeficiency Virus-Associated Neurocognitive Disorders: A Model of Ferroptosis-Induced Neurodegeneration.Front Neurosci. 2022 May 12;16:904816. doi: 10.3389/fnins.2022.904816. eCollection 2022. Front Neurosci. 2022. PMID: 35645713 Free PMC article. Review.
References
-
- Bartholomeeusen K., Xiang Y., Fujinaga K., Peterlin B. M. (2012). Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J. Biol. Chem. 287, 36609–36616. 10.1074/jbc.M112.410746 - DOI - PMC - PubMed
-
- Bliss C. (1939). The toxicity of poisons applied jointly1. Ann. Appl. Biol. 26, 585–615. 10.1111/j.1744-7348.1939.tb06990.x - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

