Mapping the allosteric sites of the A2A adenosine receptor
- PMID: 28639411
- PMCID: PMC5741531
- DOI: 10.1111/cbdd.13053
Mapping the allosteric sites of the A2A adenosine receptor
Abstract
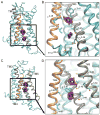
The A2A adenosine receptor (A2A AR) is a G protein-coupled receptor that is pharmacologically targeted for the treatment of inflammation, sepsis, cancer, neurodegeneration, and Parkinson's disease. Recently, we applied long-timescale molecular dynamics simulations on two ligand-free receptor conformations, starting from the agonist-bound (PDB ID: 3QAK) and antagonist-bound (PDB ID: 3EML) X-ray structures. This analysis revealed four distinct conformers of the A2A AR: the active, intermediate 1, intermediate 2, and inactive. In this study, we apply the fragment-based mapping algorithm, FTMap, on these receptor conformations to uncover five non-orthosteric sites on the A2A AR. Two sites that are identified in the active conformation are located in the intracellular region of the transmembrane helices (TM) 3/TM4 and the G protein-binding site in the intracellular region between TM2/TM3/TM6/TM7. Three sites are identified in the intermediate 1 and intermediate 2 conformations, annexing a site in the lipid interface of TM5/TM6. Five sites are identified in the inactive conformation, comprising a site in the intracellular region of TM1/TM7 and in the extracellular region of TM3/TM4 of the A2A AR. We postulate that these sites on the A2A AR be screened for allosteric modulators for the treatment of inflammatory and neurological diseases.
Keywords: GPCR; A2A adenosine receptor; FTMap; G protein-coupled receptors; allostery; fragment mapping.
© 2017 John Wiley & Sons A/S.
Conflict of interest statement
The authors declare that there are no conflicts of interests.
Figures




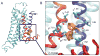
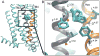
References
-
- Thiel M, Caldwell CC, Sitkovsky MV. The critical role of adenosine A2A receptors in downregulation of inflammation and immunity in the pathogenesis of infectious diseases. Microbes and Infection. 2003;5(6):515–526. - PubMed
-
- Sivak KV, et al. Adenosine A2A receptor as a drug target for treatment of sepsis. Molecular Biology. 2016;50(2):200–212. - PubMed
-
- Cekic C, Linden J. Purinergic regulation of the immune system. Nature Reviews Immunology. 2016;16(3):177–192. - PubMed
-
- Pinna A. Adenosine A2A receptor antagonists in Parkinson’s disease: progress in clinical trials from the newly approved istradefylline to drugs in early development and those already discontinued. CNS Drugs. 2014;28(5):455–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

