Boolean analysis reveals systematic interactions among low-abundance species in the human gut microbiome
- PMID: 28640804
- PMCID: PMC5480827
- DOI: 10.1371/journal.pcbi.1005361
Boolean analysis reveals systematic interactions among low-abundance species in the human gut microbiome
Abstract
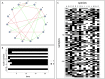
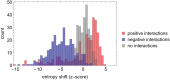
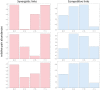
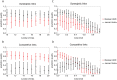
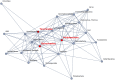
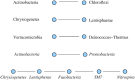
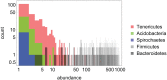
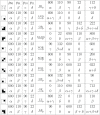
The analysis of microbiome compositions in the human gut has gained increasing interest due to the broader availability of data and functional databases and substantial progress in data analysis methods, but also due to the high relevance of the microbiome in human health and disease. While most analyses infer interactions among highly abundant species, the large number of low-abundance species has received less attention. Here we present a novel analysis method based on Boolean operations applied to microbial co-occurrence patterns. We calibrate our approach with simulated data based on a dynamical Boolean network model from which we interpret the statistics of attractor states as a theoretical proxy for microbiome composition. We show that for given fractions of synergistic and competitive interactions in the model our Boolean abundance analysis can reliably detect these interactions. Analyzing a novel data set of 822 microbiome compositions of the human gut, we find a large number of highly significant synergistic interactions among these low-abundance species, forming a connected network, and a few isolated competitive interactions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nature Biotechnology. 2013;31(9):814–821. 10.1038/nbt.2676 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

