Whole genome analysis of selected human and animal rotaviruses identified in Uganda from 2012 to 2014 reveals complex genome reassortment events between human, bovine, caprine and porcine strains
- PMID: 28640820
- PMCID: PMC5480867
- DOI: 10.1371/journal.pone.0178855
Whole genome analysis of selected human and animal rotaviruses identified in Uganda from 2012 to 2014 reveals complex genome reassortment events between human, bovine, caprine and porcine strains
Abstract
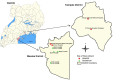
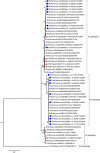
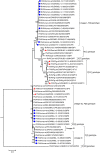
Rotaviruses of species A (RVA) are a common cause of diarrhoea in children and the young of various other mammals and birds worldwide. To investigate possible interspecies transmission of RVAs, whole genomes of 18 human and 6 domestic animal RVA strains identified in Uganda between 2012 and 2014 were sequenced using the Illumina HiSeq platform. The backbone of the human RVA strains had either a Wa- or a DS-1-like genetic constellation. One human strain was a Wa-like mono-reassortant containing a DS-1-like VP2 gene of possible animal origin. All eleven genes of one bovine RVA strain were closely related to those of human RVAs. One caprine strain had a mixed genotype backbone, suggesting that it emerged from multiple reassortment events involving different host species. The porcine RVA strains had mixed genotype backbones with possible multiple reassortant events with strains of human and bovine origin.Overall, whole genome characterisation of rotaviruses found in domestic animals in Uganda strongly suggested the presence of human-to animal RVA transmission, with concomitant circulation of multi-reassortant strains potentially derived from complex interspecies transmission events. However, whole genome data from the human RVA strains causing moderate and severe diarrhoea in under-fives in Uganda indicated that they were primarily transmitted from person-to-person.
Conflict of interest statement
Figures





References
-
- Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Archives of virology. 2012;157(6):1177–82. Epub 2012/03/21. doi: 10.1007/s00705-012-1273-3 . - DOI - PubMed
-
- Mihalov-Kovacs E, Gellert A, Marton S, Farkas SL, Feher E, Oldal M, et al. Candidate new rotavirus species in sheltered dogs, Hungary. Emerging infectious diseases. 2015;21(4):660–3. Epub 2015/03/27. doi: 10.3201/eid2104.141370 ; PubMed Central PMCID: PMCPMC4378476. - DOI - PMC - PubMed
-
- Banyai K, Kemenesi G, Budinski I, Foldes F, Zana B, Marton S, et al. Candidate new rotavirus species in Schreiber's bats, Serbia. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2016;48:19–26. Epub 2016/12/10. doi: 10.1016/j.meegid.2016.12.002 . - DOI - PMC - PubMed
-
- Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009;136(6):1939–51. Epub 2009/05/22. doi: 10.1053/j.gastro.2009.02.076 ; PubMed Central PMCID: PMCPMC3690811. - DOI - PMC - PubMed
-
- Desselberger U. Rotaviruses. Virus research. 2014;190:75–96. Epub 2014/07/13. doi: 10.1016/j.virusres.2014.06.016 . - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

