Template-switching during replication fork repair in bacteria
- PMID: 28641943
- PMCID: PMC5538584
- DOI: 10.1016/j.dnarep.2017.06.014
Template-switching during replication fork repair in bacteria
Abstract
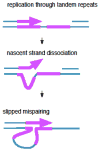
Replication forks frequently are challenged by lesions on the DNA template, replication-impeding DNA secondary structures, tightly bound proteins or nucleotide pool imbalance. Studies in bacteria have suggested that under these circumstances the fork may leave behind single-strand DNA gaps that are subsequently filled by homologous recombination, translesion DNA synthesis or template-switching repair synthesis. This review focuses on the template-switching pathways and how the mechanisms of these processes have been deduced from biochemical and genetic studies. I discuss how template-switching can contribute significantly to genetic instability, including mutational hotspots and frequent genetic rearrangements, and how template-switching may be elicited by replication fork damage.
Keywords: Copy number variation; DNA replication; Genetic recombination; Mutagenesis; Postreplication repair; Quasipalindrome.
Copyright © 2017 Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures







References
-
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
-
- Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair. 2006;5:1495–1498. - PubMed
-
- Bichara M, Meier M, Wagner J, Cordonnier A, Lambert IB. Postreplication repair mechanisms in the presence of DNA adducts in Escherichia coli. Mutation Res. 2011;727:104–122. - PubMed
-
- Heller RC, Marians KJ. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol Cell. 2005;17:733–743. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

