Mediator 1 Is Atherosclerosis Protective by Regulating Macrophage Polarization
- PMID: 28642237
- PMCID: PMC5739054
- DOI: 10.1161/ATVBAHA.117.309672
Mediator 1 Is Atherosclerosis Protective by Regulating Macrophage Polarization
Abstract
Objective: MED1 (mediator 1) interacts with transcription factors to regulate transcriptional machinery. The role of MED1 in macrophage biology and the relevant disease state remains to be investigated.
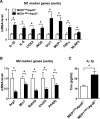
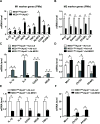
Approach and results: To study the molecular mechanism by which MED1 regulates the M1/M2 phenotype switch of macrophage and the effect on atherosclerosis, we generated MED1/apolipoprotein E (ApoE) double-deficient (MED1ΔMac/ApoE-/-) mice and found that atherosclerosis was greater in MED1ΔMac/ApoE-/- mice than in MED1fl/fl/ApoE-/- littermates. The gene expression of M1 markers was increased and that of M2 markers decreased in both aortic wall and peritoneal macrophages from MED1ΔMac/ApoE-/- mice, whereas MED1 overexpression rectified the changes in M1/M2 expression. Moreover, LDLR (low-density lipoprotein receptor)-deficient mice received bone marrow from MED1ΔMac mice showed greater atherosclerosis. Mechanistically, MED1 ablation decreased the binding of PPARγ (peroxisome proliferator-activated receptor γ) and enrichment of H3K4me1 and H3K27ac to upstream region of M2 marker genes. Furthermore, interleukin 4 induction of PPARγ and MED1 increased the binding of PPARγ or MED1 to the PPAR response elements of M2 marker genes.
Conclusions: Our data suggest that MED1 is required for the PPARγ-mediated M2 phenotype switch, with M2 marker genes induced but M1 marker genes suppressed. MED1 in macrophages has an antiatherosclerotic role via PPARγ-regulated transactivation.
Keywords: apolipoproteins; atherosclerosis; interleukins; macrophages; transcription factors.
© 2017 American Heart Association, Inc.
Figures





References
-
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. - PubMed
-
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. - PubMed
-
- Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

