Volumetric Analysis from a Harmonized Multisite Brain MRI Study of a Single Subject with Multiple Sclerosis
- PMID: 28642263
- PMCID: PMC5557658
- DOI: 10.3174/ajnr.A5254
Volumetric Analysis from a Harmonized Multisite Brain MRI Study of a Single Subject with Multiple Sclerosis
Abstract
Background and purpose: MR imaging can be used to measure structural changes in the brains of individuals with multiple sclerosis and is essential for diagnosis, longitudinal monitoring, and therapy evaluation. The North American Imaging in Multiple Sclerosis Cooperative steering committee developed a uniform high-resolution 3T MR imaging protocol relevant to the quantification of cerebral lesions and atrophy and implemented it at 7 sites across the United States. To assess intersite variability in scan data, we imaged a volunteer with relapsing-remitting MS with a scan-rescan at each site.
Materials and methods: All imaging was acquired on Siemens scanners (4 Skyra, 2 Tim Trio, and 1 Verio). Expert segmentations were manually obtained for T1-hypointense and T2 (FLAIR) hyperintense lesions. Several automated lesion-detection and whole-brain, cortical, and deep gray matter volumetric pipelines were applied. Statistical analyses were conducted to assess variability across sites, as well as systematic biases in the volumetric measurements that were site-related.
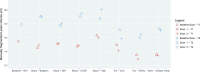
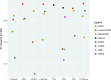
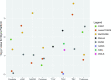
Results: Systematic biases due to site differences in expert-traced lesion measurements were significant (P < .01 for both T1 and T2 lesion volumes), with site explaining >90% of the variation (range, 13.0-16.4 mL in T1 and 15.9-20.1 mL in T2) in lesion volumes. Site also explained >80% of the variation in most automated volumetric measurements. Output measures clustered according to scanner models, with similar results from the Skyra versus the other 2 units.
Conclusions: Even in multicenter studies with consistent scanner field strength and manufacturer after protocol harmonization, systematic differences can lead to severe biases in volumetric analyses.
© 2017 by American Journal of Neuroradiology.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
