Enrichment of low-frequency functional variants revealed by whole-genome sequencing of multiple isolated European populations
- PMID: 28643794
- PMCID: PMC5490002
- DOI: 10.1038/ncomms15927
Enrichment of low-frequency functional variants revealed by whole-genome sequencing of multiple isolated European populations
Abstract
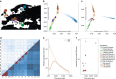
The genetic features of isolated populations can boost power in complex-trait association studies, and an in-depth understanding of how their genetic variation has been shaped by their demographic history can help leverage these advantageous characteristics. Here, we perform a comprehensive investigation using 3,059 newly generated low-depth whole-genome sequences from eight European isolates and two matched general populations, together with published data from the 1000 Genomes Project and UK10K. Sequencing data give deeper and richer insights into population demography and genetic characteristics than genotype-chip data, distinguishing related populations more effectively and allowing their functional variants to be studied more fully. We demonstrate relaxation of purifying selection in the isolates, leading to enrichment of rare and low-frequency functional variants, using novel statistics, DVxy and SVxy. We also develop an isolation-index (Isx) that predicts the overall level of such key genetic characteristics and can thus help guide population choice in future complex-trait association studies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

