The genomic landscape of tuberous sclerosis complex
- PMID: 28643795
- PMCID: PMC5481739
- DOI: 10.1038/ncomms15816
The genomic landscape of tuberous sclerosis complex
Abstract
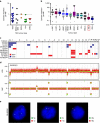
Tuberous sclerosis complex (TSC) is a rare genetic disease causing multisystem growth of benign tumours and other hamartomatous lesions, which leads to diverse and debilitating clinical symptoms. Patients are born with TSC1 or TSC2 mutations, and somatic inactivation of wild-type alleles drives MTOR activation; however, second hits to TSC1/TSC2 are not always observed. Here, we present the genomic landscape of TSC hamartomas. We determine that TSC lesions contain a low somatic mutational burden relative to carcinomas, a subset feature large-scale chromosomal aberrations, and highly conserved molecular signatures for each type exist. Analysis of the molecular signatures coupled with computational approaches reveals unique aspects of cellular heterogeneity and cell origin. Using immune data sets, we identify significant neuroinflammation in TSC-associated brain tumours. Taken together, this molecular catalogue of TSC serves as a resource into the origin of these hamartomas and provides a framework that unifies genomic and transcriptomic dimensions for complex tumours.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Osborne J. P., Fryer A. & Webb D. Epidemiology of tuberous sclerosis. Ann. N. Y. Acad. Sci. 615, 125–127 (1991). - PubMed
-
- European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 75, 1305–1315 (1993). - PubMed
-
- Gomez M. R. Criteria for diagnosis. in Tuberous Sclerosis ed. Gomez M. R. 10–23Raven Press (1988).
-
- Roach E. S., Gomez M. R. & Northrup H. Tuberous sclerosis complex consensus conference: revised clinical diagnostic criteria. J. Child Neurol. 13, 624–628 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

