Lytic transglycosylases: concinnity in concision of the bacterial cell wall
- PMID: 28644060
- PMCID: PMC6102726
- DOI: 10.1080/10409238.2017.1337705
Lytic transglycosylases: concinnity in concision of the bacterial cell wall
Abstract
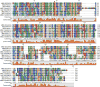
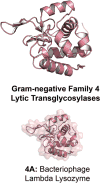
The lytic transglycosylases (LTs) are bacterial enzymes that catalyze the non-hydrolytic cleavage of the peptidoglycan structures of the bacterial cell wall. They are not catalysts of glycan synthesis as might be surmised from their name. Notwithstanding the seemingly mundane reaction catalyzed by the LTs, their lytic reactions serve bacteria for a series of astonishingly diverse purposes. These purposes include cell-wall synthesis, remodeling, and degradation; for the detection of cell-wall-acting antibiotics; for the expression of the mechanism of cell-wall-acting antibiotics; for the insertion of secretion systems and flagellar assemblies into the cell wall; as a virulence mechanism during infection by certain Gram-negative bacteria; and in the sporulation and germination of Gram-positive spores. Significant advances in the mechanistic understanding of each of these processes have coincided with the successive discovery of new LTs structures. In this review, we provide a systematic perspective on what is known on the structure-function correlations for the LTs, while simultaneously identifying numerous opportunities for the future study of these enigmatic enzymes.
Keywords: AmpC; AmpR; Lytic transglycosylase; bacteria; cell-wall recycling; muropeptide; peptidoglycan; secretion system.
Conflict of interest statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article. This work was supported by a grant to S.M. from the National Institute of Health (NIH Grant GM61629). D.A.D. is a Fellow of the Chemistry-Biochemistry-Biology Interface (CBBI) Program (NIH Training Grant T32GM075762) and a Fellow of the ECK Institute of Global Health at the University of Notre Dame.
Figures
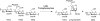









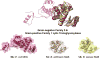



References
-
- Alcorlo M, Martínez-Caballero S, Molina R, et al. Carbohydrate recognition and lysis by bacterial peptidoglycan hydrolases. Curr Opin Struct Biol. 2017;44:87–100. - PubMed
-
- Alvarez L, Hernandez SB, de Pedro MA, et al. Ultra-sensitive, high-resolution liquid chromatography methods for the high-throughput quantitative analysis of bacterial cell wall chemistry and structure. Methods Mol Biol. 2016;1440:11–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
