ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline
- PMID: 28645154
- PMCID: PMC5587817
- DOI: 10.1093/nar/gkx536
ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline
Abstract
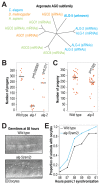
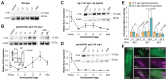
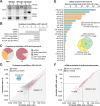
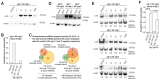
Caenorhabditis elegans contains 25 Argonautes, of which, ALG-1 and ALG-2 are known to primarily interact with miRNAs. ALG-5 belongs to the AGO subfamily of Argonautes that includes ALG-1 and ALG-2, but its role in small RNA pathways is unknown. We analyzed by high-throughput sequencing the small RNAs associated with ALG-5, ALG-1 and ALG-2, as well as changes in mRNA expression in alg-5, alg-1 and alg-2 mutants. We show that ALG-5 defines a distinct branch of the miRNA pathway affecting the expression of genes involved in immunity, defense, and development. In contrast to ALG-1 and ALG-2, which associate with most miRNAs and have general roles throughout development, ALG-5 interacts with only a small subset of miRNAs and is specifically expressed in the germline where it localizes alongside the piRNA and siRNA machinery at P granules. alg-5 is required for optimal fertility and mutations in alg-5 lead to a precocious transition from spermatogenesis to oogenesis. Our results provide a near-comprehensive analysis of miRNA-Argonaute interactions in C. elegans and reveal a new role for miRNAs in the germline.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. - PubMed
-
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003; 425:415–419. - PubMed
-
- Khvorova A., Reynolds A., Jayasena S.D.. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003; 115:209–216. - PubMed
-
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D.. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003; 115:199–208. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

