Transcriptomic and Proteomic Profiling Provides Insight into Mesangial Cell Function in IgA Nephropathy
- PMID: 28646076
- PMCID: PMC5619958
- DOI: 10.1681/ASN.2016101103
Transcriptomic and Proteomic Profiling Provides Insight into Mesangial Cell Function in IgA Nephropathy
Abstract
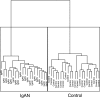
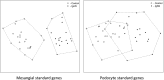
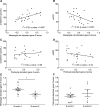
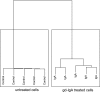
IgA nephropathy (IgAN), the most common GN worldwide, is characterized by circulating galactose-deficient IgA (gd-IgA) that forms immune complexes. The immune complexes are deposited in the glomerular mesangium, leading to inflammation and loss of renal function, but the complete pathophysiology of the disease is not understood. Using an integrated global transcriptomic and proteomic profiling approach, we investigated the role of the mesangium in the onset and progression of IgAN. Global gene expression was investigated by microarray analysis of the glomerular compartment of renal biopsy specimens from patients with IgAN (n=19) and controls (n=22). Using curated glomerular cell type-specific genes from the published literature, we found differential expression of a much higher percentage of mesangial cell-positive standard genes than podocyte-positive standard genes in IgAN. Principal coordinate analysis of expression data revealed clear separation of patient and control samples on the basis of mesangial but not podocyte cell-positive standard genes. Additionally, patient clinical parameters (serum creatinine values and eGFRs) significantly correlated with Z scores derived from the expression profile of mesangial cell-positive standard genes. Among patients grouped according to Oxford MEST score, patients with segmental glomerulosclerosis had a significantly higher mesangial cell-positive standard gene Z score than patients without segmental glomerulosclerosis. By investigating mesangial cell proteomics and glomerular transcriptomics, we identified 22 common pathways induced in mesangial cells by gd-IgA, most of which mediate inflammation. The genes, proteins, and corresponding pathways identified provide novel insights into the pathophysiologic mechanisms leading to IgAN.
Keywords: IgA nephropathy; mesangial cells; transcriptional profiling.
Copyright © 2017 by the American Society of Nephrology.
Figures
References
-
- Cove-Smith A, Hendry BM: The regulation of mesangial cell proliferation. Nephron, Exp Nephrol 108: e74–e79, 2008 - PubMed
-
- Hendry BM, Khwaja A, Qu QY, Shankland SJ: Distinct functions for Ras GTPases in the control of proliferation and apoptosis in mouse and human mesangial cells. Kidney Int 69: 99–104, 2006 - PubMed
-
- Cattran DC, Coppo R, Cook HT, Feehally J, Roberts IS, Troyanov S, Alpers CE, Amore A, Barratt J, Berthoux F, Bonsib S, Bruijn JA, D’Agati V, D’Amico G, Emancipator S, Emma F, Ferrario F, Fervenza FC, Florquin S, Fogo A, Geddes CC, Groene HJ, Haas M, Herzenberg AM, Hill PA, Hogg RJ, Hsu SI, Jennette JC, Joh K, Julian BA, Kawamura T, Lai FM, Leung CB, Li LS, Li PK, Liu ZH, Mackinnon B, Mezzano S, Schena FP, Tomino Y, Walker PD, Wang H, Weening JJ, Yoshikawa N, Zhang H; Working Group of the International IgA Nephropathy Network and the Renal Pathology Society : The oxford classification of IgA nephropathy: Rationale, clinicopathological correlations, and classification. Kidney Int 76: 534–545, 2009 - PubMed
-
- Moresco RN, Speeckaert MM, Delanghe JR: Diagnosis and monitoring of IgA nephropathy: The role of biomarkers as an alternative to renal biopsy. Autoimmun Rev 14: 847–853, 2015 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

