Vaccine candidate discovery for the next generation of malaria vaccines
- PMID: 28646586
- PMCID: PMC5588761
- DOI: 10.1111/imm.12780
Vaccine candidate discovery for the next generation of malaria vaccines
Abstract
Although epidemiological observations, IgG passive transfer studies and experimental infections in humans all support the feasibility of developing highly effective malaria vaccines, the precise antigens that induce protective immunity remain uncertain. Here, we review the methodologies applied to vaccine candidate discovery for Plasmodium falciparum malaria from the pre- to post-genomic era. Probing of genomic and cDNA libraries with antibodies of defined specificities or functional activity predominated the former, whereas reverse vaccinology encompassing high throughput in silico analyses of genomic, transcriptomic or proteomic parasite data sets is the mainstay of the latter. Antibody-guided vaccine design spanned both eras but currently benefits from technological advances facilitating high-throughput screening and downstream applications. We make the case that although we have exponentially increased our ability to identify numerous potential vaccine candidates in a relatively short space of time, a significant bottleneck remains in their validation and prioritization for evaluation in clinical trials. Longitudinal cohort studies provide supportive evidence but results are often conflicting between studies. Demonstration of antigen-specific antibody function is valuable but the relative importance of one mechanism over another with regards to protection remains undetermined. Animal models offer useful insights but may not accurately reflect human disease. Challenge studies in humans are preferable but prohibitively expensive. In the absence of reliable correlates of protection, suitable animal models or a better understanding of the mechanisms underlying protective immunity in humans, vaccine candidate discovery per se may not be sufficient to provide the paradigm shift necessary to develop the next generation of highly effective subunit malaria vaccines.
Keywords: Plasmodium falciparum; antibodies; bioinformatics; vaccines.
© 2017 The Authors. Immunology Published by John Wiley & Sons Ltd.
Figures

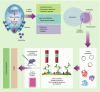
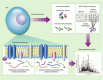
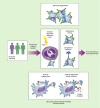
References
-
- WHO , Chan M. 2016. World Malaria report.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

