Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling
- PMID: 28648364
- PMCID: PMC5531599
- DOI: 10.1016/j.stem.2017.05.020
Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling
Erratum in
-
Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling.Cell Stem Cell. 2019 May 2;24(5):829. doi: 10.1016/j.stem.2019.04.002. Cell Stem Cell. 2019. PMID: 31051135 Free PMC article. No abstract available.
Abstract
Gastric and small intestinal organoids differentiated from human pluripotent stem cells (hPSCs) have revolutionized the study of gastrointestinal development and disease. Distal gut tissues such as cecum and colon, however, have proved considerably more challenging to derive in vitro. Here we report the differentiation of human colonic organoids (HCOs) from hPSCs. We found that BMP signaling is required to establish a posterior SATB2+ domain in developing and postnatal intestinal epithelium. Brief activation of BMP signaling is sufficient to activate a posterior HOX code and direct hPSC-derived gut tube cultures into HCOs. In vitro, HCOs express colonic markers and contained colon-specific cell populations. Following transplantation into mice, HCOs undergo morphogenesis and maturation to form tissue that exhibits molecular, cellular, and morphologic properties of human colon. Together these data show BMP-dependent patterning of human hindgut into HCOs, which will be valuable for studying diseases including colitis and colon cancer.
Keywords: BMP signaling; Satb2; colonic organoids; hindgut; posterior HOX code.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
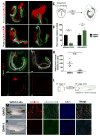
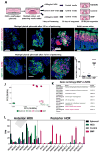

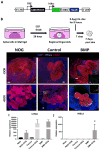

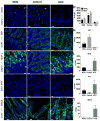
Comment in
-
Stem cells: Colonic organoids for drug testing and colorectal disease modelling.Nat Rev Mol Cell Biol. 2017 Aug;18(8):467. doi: 10.1038/nrm.2017.70. Epub 2017 Jul 5. Nat Rev Mol Cell Biol. 2017. PMID: 28676698 No abstract available.
-
Organoids recapitulate organs?Stem Cell Investig. 2018 Jan 17;5:3. doi: 10.21037/sci.2018.01.02. eCollection 2018. Stem Cell Investig. 2018. PMID: 29430459 Free PMC article. No abstract available.
References
-
- Beuling E, Bosse T, Buckner MA, Krasinski SD. Co-localization of Gata4 and Hnf1 alpha in the gastrointestinal tract is restricted to the distal stomach and proximal small intestine. Gastroenterology. 2007a;132:A586–A586.
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK070858/DK/NIDDK NIH HHS/United States
- R01 DK102551/DK/NIDDK NIH HHS/United States
- P30 DK078392/DK/NIDDK NIH HHS/United States
- U24 DK085532/DK/NIDDK NIH HHS/United States
- U01 DK103141/DK/NIDDK NIH HHS/United States
- R01 DK092456/DK/NIDDK NIH HHS/United States
- U19 AI116491/AI/NIAID NIH HHS/United States
- R01 DK098350/DK/NIDDK NIH HHS/United States
- U01 DK103117/DK/NIDDK NIH HHS/United States
- P01 HD093363/HD/NICHD NIH HHS/United States
- U18 EB021780/EB/NIBIB NIH HHS/United States
- U01 DK085532/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

