Targeted Protein Degradation: from Chemical Biology to Drug Discovery
- PMID: 28648379
- PMCID: PMC5610075
- DOI: 10.1016/j.chembiol.2017.05.024
Targeted Protein Degradation: from Chemical Biology to Drug Discovery
Abstract
Traditional pharmaceutical drug discovery is almost exclusively focused on directly controlling protein activity to cure diseases. Modulators of protein activity, especially inhibitors, are developed and applied at high concentration to achieve maximal effects. Thereby, reduced bioavailability and off-target effects can hamper compound efficacy. Nucleic acid-based strategies that control protein function by affecting expression have emerged as an alternative. However, metabolic stability and broad bioavailability represent development hurdles that remain to be overcome for these approaches. More recently, utilizing the cell's own protein destruction machinery for selective degradation of essential drivers of human disorders has opened up a new and exciting area of drug discovery. Small-molecule-induced proteolysis of selected substrates offers the potential of reaching beyond the limitations of the current pharmaceutical paradigm to expand the druggable target space.
Keywords: E3 ligase; PROTACs; hydrophobic tagging; protein degradation.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

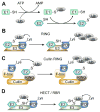
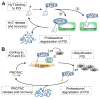
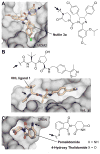
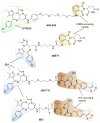

References
-
- Adjei AA. What is the right dose? The elusive optimal biologic dose in phase I clinical trials. J Clin Oncol. 2006;24:4054–4055. - PubMed
-
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. - PubMed
-
- Bielskiene K, Bagdoniene L, Mozuraitiene J, Kazbariene B, Janulionis E. E3 ubiquitin ligases as drug targets and prognostic biomarkers in melanoma. Medicina (Kaunas) 2015;51:1–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

