The structural requirements of histone deacetylase inhibitors: SAHA analogs modified at the C5 position display dual HDAC6/8 selectivity
- PMID: 28648461
- PMCID: PMC5747257
- DOI: 10.1016/j.bmcl.2017.06.033
The structural requirements of histone deacetylase inhibitors: SAHA analogs modified at the C5 position display dual HDAC6/8 selectivity
Abstract
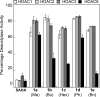
Histone deacetylase (HDAC) proteins have emerged as important targets for anti-cancer drugs, with four small molecules approved for use in the clinic. Suberoylanilide hydroxamic acid (Vorinostat, SAHA) was the first FDA-approved HDAC inhibitor for cancer treatment. However, SAHA inhibits most of the eleven HDAC isoforms. To understand the structural requirements of HDAC inhibitor selectivity and develop isoform selective HDAC inhibitors, SAHA analogs modified in the linker at the C5 position were synthesized and tested for potency and selectivity. C5-modified SAHA analogs displayed dual selectivity to HDAC6 and HDAC8 over HDAC 1, 2, and 3, with only a modest reduction in potency. These findings are consistent with prior work showing that modification of the linker region of SAHA can alter isoform selectivity. The observed HDAC6/8 selectivity of C5-modified SAHA analogs provide guidance toward development of isoform selective HDAC inhibitors and more effective anti-cancer drugs.
Keywords: HDAC inhibitors; HDAC6/8 selective inhibitor; Histone deacetylase; Isoform selectivity; Vorinostat.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures




References
-
- Walsh CT, Garneau-Tsodikova S, Gatto GJ. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem, Int Ed. 2005;44(45):7342–7372. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

