A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus
- PMID: 28648617
- PMCID: PMC5573227
- DOI: 10.1016/j.jmb.2017.06.015
A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus
Abstract
Influenza virus evolves rapidly to constantly escape from natural immunity. Most humoral immune responses to influenza virus target the hemagglutinin (HA) glycoprotein, which is the major antigen on the surface of the virus. The HA is composed of a globular head domain for receptor binding and a stem domain for membrane fusion. The major antigenic sites of HA are located in the globular head subdomain, which is highly tolerant of amino acid substitutions and continual addition of glycosylation sites. Nonetheless, the evolution of the receptor-binding site and the stem region on HA is severely constrained by their functional roles in engaging the host receptor and in mediating membrane fusion, respectively. Here, we review how broadly neutralizing antibodies (bnAbs) exploit these evolutionary constraints to protect against diverse influenza strains. We also discuss the emerging role of other epitopes that are conserved only in subsets of viruses. This rapidly increasing knowledge of the evolutionary biology, immunology, structural biology, and virology of influenza virus is invaluable for development and design of more universal influenza vaccines and novel therapeutics.
Keywords: antibody; evolution; hemagglutinin; influenza A virus; protein structure.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures


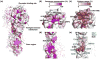

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

