NTL8 Regulates Trichome Formation in Arabidopsis by Directly Activating R3 MYB Genes TRY and TCL1
- PMID: 28649093
- PMCID: PMC5543959
- DOI: 10.1104/pp.17.00510
NTL8 Regulates Trichome Formation in Arabidopsis by Directly Activating R3 MYB Genes TRY and TCL1
Abstract
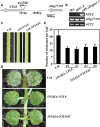
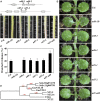
The NAM, ATAF1/2, and CUC (NAC) are plant-specific transcription factors that regulate multiple aspects of plant growth and development and plant response to environmental stimuli. We report here the identification of NTM1-LIKE8 (NTL8), a membrane-associated NAC transcription factor, as a novel regulator of trichome formation in Arabidopsis (Arabidopsis thaliana). From an activation-tagged Arabidopsis population, we identified a dominant, gain-of-function mutant with glabrous inflorescence stem. By using plasmid rescue and RT-PCR analyses, we found that NTL8 was tagged; thus, the mutant was named ntl8-1 Dominant (ntl8-1D). Recapitulation experiment further confirmed that the phenotype observed in the ntl8-1D mutant was caused by elevated expression of NTL8 Quantitative RT-PCR results showed that the expression level of the single-repeat R3 MYB genes TRIPTYCHON (TRY) and TRICHOMELESS1 (TCL1) was elevated in the ntl8-1D mutant. Genetic analyses demonstrated that NTL8 acts upstream of TRY and TCL1 in the regulation of trichome formation. When recruited to the promoter region of the reporter gene Gal4:GUS by a fused GAL4 DNA-binding domain, NTL8 activated the expression of the reporter gene. Chromatin immunoprecipitation results indicated that TRY and TCL1 are direct targets of NTL8. However, NTL8 did not interact with SQUAMOSA PROMOTER BINDING PROTEIN LIKE9, another transcription factor that regulates the expression of TRY and TCL1, in yeast and plant cells. Taken together, our results suggest that NTL8 negatively regulates trichome formation in Arabidopsis by directly activating the expression of TRY and TCL1.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures





Similar articles
-
Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in Arabidopsis.BMC Plant Biol. 2018 Apr 13;18(1):63. doi: 10.1186/s12870-018-1271-z. BMC Plant Biol. 2018. PMID: 29653514 Free PMC article.
-
Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis.BMC Plant Biol. 2011 Dec 15;11:176. doi: 10.1186/1471-2229-11-176. BMC Plant Biol. 2011. PMID: 22168948 Free PMC article.
-
TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis.Development. 2007 Nov;134(21):3873-82. doi: 10.1242/dev.009597. Development. 2007. PMID: 17933793
-
Regulation of root hair cell differentiation by R3 MYB transcription factors in tomato and Arabidopsis.Front Plant Sci. 2014 Mar 13;5:91. doi: 10.3389/fpls.2014.00091. eCollection 2014. Front Plant Sci. 2014. PMID: 24659995 Free PMC article. Review.
-
Progress on trichome development regulated by phytohormone signaling.Plant Signal Behav. 2011 Dec;6(12):1959-62. doi: 10.4161/psb.6.12.18120. Plant Signal Behav. 2011. PMID: 22105030 Free PMC article. Review.
Cited by
-
The Carboxyl-Terminus of TRANSPARENT TESTA GLABRA1 Is Critical for Its Functions in Arabidopsis.Int J Mol Sci. 2021 Sep 17;22(18):10039. doi: 10.3390/ijms221810039. Int J Mol Sci. 2021. PMID: 34576199 Free PMC article.
-
Involvement of ABA Responsive SVB Genes in the Regulation of Trichome Formation in Arabidopsis.Int J Mol Sci. 2021 Jun 24;22(13):6790. doi: 10.3390/ijms22136790. Int J Mol Sci. 2021. PMID: 34202673 Free PMC article.
-
AtbZIP62 Acts as a Transcription Repressor to Positively Regulate ABA Responses in Arabidopsis.Plants (Basel). 2022 Nov 10;11(22):3037. doi: 10.3390/plants11223037. Plants (Basel). 2022. PMID: 36432766 Free PMC article.
-
Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in Arabidopsis.BMC Plant Biol. 2018 Apr 13;18(1):63. doi: 10.1186/s12870-018-1271-z. BMC Plant Biol. 2018. PMID: 29653514 Free PMC article.
-
TCP transcription factors suppress cotyledon trichomes by impeding a cell differentiation-regulating complex.Plant Physiol. 2021 May 27;186(1):434-451. doi: 10.1093/plphys/kiab053. Plant Physiol. 2021. PMID: 33576799 Free PMC article.
References
-
- An L, Zhou Z, Sun L, Yan A, Xi W, Yu N, Cai W, Chen X, Yu H, Schiefelbein J, et al. (2012) A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. Plant J 72: 474–490 - PubMed
-
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
-
- Dai X, Zhou L, Zhang W, Cai L, Guo H, Tian H, Schiefelbein J, Wang S (2016) A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ 39: 897–907 - PubMed
-
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

