Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity
- PMID: 28649403
- PMCID: PMC5445578
- DOI: 10.1038/s41522-016-0011-0
Inter-personal diversity and temporal dynamics of dental, tongue, and salivary microbiota in the healthy oral cavity
Abstract
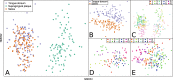
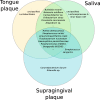
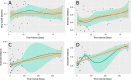
Oral microbes form a complex and dynamic biofilm community, which is subjected to daily host and environmental challenges. Dysbiosis of the oral biofilm is correlated with local and distal infections and postulating a baseline for the healthy core oral microbiota provides an opportunity to examine such shifts during the onset and recurrence of disease. Here we quantified the daily, weekly, and monthly variability of the oral microbiome by sequencing the largest oral microbiota time-series to date, covering multiple oral sites in ten healthy individuals. Temporal dynamics of salivary, dental, and tongue consortia were examined by high-throughput 16S rRNA gene sequencing over 90 days, with four individuals sampled additionally 1 year later. Distinct communities were observed between dental, tongue, and salivary samples, with high levels of similarity observed between the tongue and salivary communities. Twenty-six core OTUs that classified within Streptococcus, Fusobacterium, Haemophilus, Neisseria, Prevotella, and Rothia genera were present in ≥95% samples and accounted for ~65% of the total sequence data. Phylogenetic diversity varied from person to person, but remained relatively stable within individuals over time compared to inter-individual variation. In contrast, the composition of rare microorganisms was highly variable over time, within most individuals. Using machine learning, an individual's oral microbial assemblage could be correctly assigned to them with 88-97% accuracy, depending on the sample site; 83% of samples taken a year after initial sampling could be confidently traced back to the source subject.
Figures

References
-
- Marsh, P. D., Martin, M. V., Lewis, M. A. & Williams, D. Oral Microbiol. (Elsevier Health Sciences, 2009).
LinkOut - more resources
Full Text Sources
Other Literature Sources

