Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module
- PMID: 28649719
- PMCID: PMC5542871
- DOI: 10.1111/nph.14638
Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module
Abstract
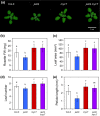
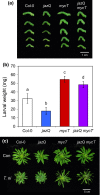
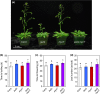
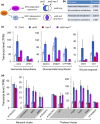
The plant hormone jasmonate (JA) promotes the degradation of JASMONATE ZIM-DOMAIN (JAZ) proteins to relieve repression on diverse transcription factors (TFs) that execute JA responses. However, little is known about how combinatorial complexity among JAZ-TF interactions maintains control over myriad aspects of growth, development, reproduction, and immunity. We used loss-of-function mutations to define epistatic interactions within the core JA signaling pathway and to investigate the contribution of MYC TFs to JA responses in Arabidopsis thaliana. Constitutive JA signaling in a jaz quintuple mutant (jazQ) was largely eliminated by mutations that block JA synthesis or perception. Comparison of jazQ and a jazQ myc2 myc3 myc4 octuple mutant validated known functions of MYC2/3/4 in root growth, chlorophyll degradation, and susceptibility to the pathogen Pseudomonas syringae. We found that MYC TFs also control both the enhanced resistance of jazQ leaves to insect herbivory and restricted leaf growth of jazQ. Epistatic transcriptional profiles mirrored these phenotypes and further showed that triterpenoid biosynthetic and glucosinolate catabolic genes are up-regulated in jazQ independently of MYC TFs. Our study highlights the utility of genetic epistasis to unravel the complexities of JAZ-TF interactions and demonstrates that MYC TFs exert master control over a JAZ-repressible transcriptional hierarchy that governs growth-defense balance.
Keywords: gene cluster; glucosinolate; growth-defense tradeoffs; jasmonate (JA); plant defense; plant hormone; plant-insect interaction; triterpenoid.
© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
Figures




Comment in
-
A plant's balance of growth and defense - revisited.New Phytol. 2017 Sep;215(4):1291-1294. doi: 10.1111/nph.14720. New Phytol. 2017. PMID: 28771818 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

