Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma
- PMID: 28649789
- PMCID: PMC5939569
- DOI: 10.1111/pcmr.12611
Beyond MITF: Multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma
Abstract
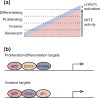
MITF governs multiple steps in the development of melanocytes, including specification from neural crest, growth, survival, and terminal differentiation. In addition, the level of MITF activity determines the phenotype adopted by melanoma cells, whether invasive, proliferative, or differentiated. However, MITF does not act alone. Here, we review literature on the transcription factors that co-regulate MITF-dependent genes. ChIP-seq studies have indicated that the transcription factors SOX10, YY1, and TFAP2A co-occupy subsets of regulatory elements bound by MITF in melanocytes. Analyses at single loci also support roles for LEF1, RB1, IRF4, and PAX3 acting in combination with MITF, while sequence motif analyses suggest that additional transcription factors colocalize with MITF at many melanocyte-specific regulatory elements. However, the precise biochemical functions of each of these MITF collaborators and their contributions to gene expression remain to be elucidated. Analogous to the transcriptional networks in morphogen-patterned tissues during embryogenesis, we anticipate that the level of MITF activity is controlled not only by the concentration of activated MITF, but also by additional transcription factors that either quantitatively or qualitatively influence the expression of MITF-target genes.
Keywords: MITF; SOX10; TFAP2A; YY1; melanocytes.
© 2017 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures



References
-
- Baldi A, Santini D, Battista T, Dragonetti E, Ferranti G, Petitti T, Paggi MG. Expression of AP-2 transcription factor and of its downstream target genes c-kit, E-cadherin and p21 in human cutaneous melanoma. Journal of Cellular Biochemistry. 2001;83:364–372. - PubMed
-
- Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development. 2004;131:1463–1477. - PubMed
-
- Begon DY, Delacroix L, Vernimmen D, Jackers P, Winkler R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. The Journal of Biological Chemistry. 2005;280:24428–24434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

