Preexisting endothelial cells mediate cardiac neovascularization after injury
- PMID: 28650345
- PMCID: PMC5531398
- DOI: 10.1172/JCI93868
Preexisting endothelial cells mediate cardiac neovascularization after injury
Abstract
The mechanisms that promote the generation of new coronary vasculature during cardiac homeostasis and after injury remain a fundamental and clinically important area of study in the cardiovascular field. Recently, it was reported that mesenchymal-to-endothelial transition (MEndoT) contributes to substantial numbers of coronary endothelial cells after myocardial infarction. Therefore, the MEndoT has been proposed as a paradigm mediating neovascularization and is considered a promising therapeutic target in cardiac regeneration. Here, we show that preexisting endothelial cells mainly beget new coronary vessels in the adult mouse heart, with essentially no contribution from other cell sources through cell-lineage transdifferentiation. Genetic-lineage tracing revealed that cardiac fibroblasts expand substantially after injury, but do not contribute to the formation of new coronary blood vessels, indicating no contribution of MEndoT to neovascularization. Moreover, genetic-lineage tracing with a pulse-chase labeling strategy also showed that essentially all new coronary vessels in the injured heart are derived from preexisting endothelial cells, but not from other cell lineages. These data indicate that therapeutic strategies for inducing neovascularization should not be based on targeting presumptive lineage transdifferentiation such as MEndoT. Instead, preexisting endothelial cells appear more likely to be the therapeutic target for promoting neovascularization and driving heart regeneration after injury.
Conflict of interest statement
Figures
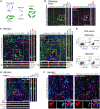
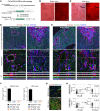


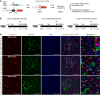


Comment in
- The relationship between cardiac endothelium and fibroblasts: it’s complicated
References
-
- Lin YD, et al. Instructive nanofiber scaffolds with VEGF create a microenvironment for arteriogenesis and cardiac repair. Sci Transl Med. 2012;4(146):146ra109. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

