Ror2-mediated alternative Wnt signaling regulates cell fate and adhesion during mammary tumor progression
- PMID: 28650466
- PMCID: PMC5658260
- DOI: 10.1038/onc.2017.206
Ror2-mediated alternative Wnt signaling regulates cell fate and adhesion during mammary tumor progression
Abstract
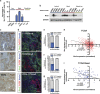
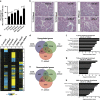
Cellular heterogeneity is a common feature in breast cancer, yet an understanding of the coexistence and regulation of various tumor cell subpopulations remains a significant challenge in cancer biology. In the current study, we approached tumor cell heterogeneity from the perspective of Wnt pathway biology to address how different modes of Wnt signaling shape the behaviors of diverse cell populations within a heterogeneous tumor landscape. Using a syngeneic TP53-null mouse model of breast cancer, we identified distinctions in the topology of canonical Wnt β-catenin-dependent signaling activity and non-canonical β-catenin-independent Ror2-mediated Wnt signaling across subtypes and within tumor cell subpopulations in vivo. We further discovered an antagonistic role for Ror2 in regulating canonical Wnt/β-catenin activity in vivo, where lentiviral shRNA depletion of Ror2 expression augmented canonical Wnt/β-catenin signaling activity across multiple basal-like models. Depletion of Ror2 expression yielded distinct phenotypic outcomes and divergent alterations in gene expression programs among different tumors, despite all sharing basal-like features. Notably, we uncovered cell state plasticity and adhesion dynamics regulated by Ror2, which influenced Ras Homology Family Member A (RhoA) and Rho-Associated Coiled-Coil Kinase 1 (ROCK1) activity downstream of Dishevelled-2 (Dvl2). Collectively, these studies illustrate the integration and collaboration of Wnt pathways in basal-like breast cancer, where Ror2 provides a spatiotemporal function to regulate the balance of Wnt signaling and cellular heterogeneity during tumor progression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

