Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles
- PMID: 28650991
- PMCID: PMC5507469
- DOI: 10.1371/journal.pgen.1006862
Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA-A, -B and -C alleles
Abstract
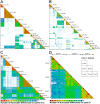
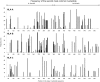
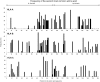
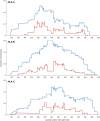
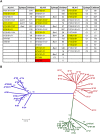
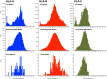
HLA class I glycoproteins contain the functional sites that bind peptide antigens and engage lymphocyte receptors. Recently, clinical application of sequence-based HLA typing has uncovered an unprecedented number of novel HLA class I alleles. Here we define the nature and extent of the variation in 3,489 HLA-A, 4,356 HLA-B and 3,111 HLA-C alleles. This analysis required development of suites of methods, having general applicability, for comparing and analyzing large numbers of homologous sequences. At least three amino-acid substitutions are present at every position in the polymorphic α1 and α2 domains of HLA-A, -B and -C. A minority of positions have an incidence >1% for the 'second' most frequent nucleotide, comprising 70 positions in HLA-A, 85 in HLA-B and 54 in HLA-C. The majority of these positions have three or four alternative nucleotides. These positions were subject to positive selection and correspond to binding sites for peptides and receptors. Most alleles of HLA class I (>80%) are very rare, often identified in one person or family, and they differ by point mutation from older, more common alleles. These alleles with single nucleotide polymorphisms reflect the germ-line mutation rate. Their frequency predicts the human population harbors 8-9 million HLA class I variants. The common alleles of human populations comprise 42 core alleles, which represent all selected polymorphism, and recombinants that have assorted this polymorphism.
Conflict of interest statement
The authors have declared that no competing interests exist. The authors NC and SYY are employed by Histogenetics Inc. There are no patents, products in development or marketed products to declare. This does not alter their adherence to all the PLOS Genetics policies on sharing data and materials, as detailed online in the guide for authors.
Figures


References
-
- Altmann DM, Trowsdale J. Major histocompatibility complex structure and function. Curr Opin Immunol. 1989;2(1):93–8. Epub 1989/10/01. - PubMed
-
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5(12):889–99. Epub 2004/12/02. doi: 10.1038/nrg1489 - DOI - PubMed
-
- Lienert K, Parham P. Evolution of MHC class I genes in higher primates. Immunol Cell Biol. 1996;74(4):349–56. doi: 10.1038/icb.1996.62 - DOI - PubMed
-
- Hughes AL, Yeager M, Carrington M. Peptide binding function and the paradox of HLA disease associations. Immunol Cell Biol. 1996;74(5):444–8. doi: 10.1038/icb.1996.74 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

