Cytomegalovirus UL128 homolog mutants that form a pentameric complex produce virus with impaired epithelial and trophoblast cell tropism and altered pathogenicity in the guinea pig
- PMID: 28651121
- PMCID: PMC5739588
- DOI: 10.1016/j.virol.2017.06.008
Cytomegalovirus UL128 homolog mutants that form a pentameric complex produce virus with impaired epithelial and trophoblast cell tropism and altered pathogenicity in the guinea pig
Abstract
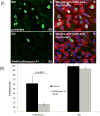
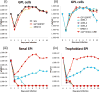
Guinea pig cytomegalovirus (GPCMV) encodes a homolog pentameric complex (PC) for specific cell tropism and congenital infection. In human cytomegalovirus, the PC is an important antibody neutralizing target and GPCMV studies will aid in the development of intervention strategies. Deletion mutants of the C-terminal domains of unique PC proteins (UL128, UL130 and UL131 homologs) were unable to form a PC in separate transient expression assays. Minor modifications to the UL128 homolog (GP129) C-terminal domain enabled PC formation but viruses encoding these mutants had altered tropism to renal and placental trophoblast cells. Mutation of the presumptive CC chemokine motif encoded by GP129 was investigated by alanine substitution of the CC motif (codons 26-27) and cysteines (codons 47 and 62). GP129 chemokine mutants formed PC but GP129 chemokine mutant viruses had reduced epitropism. A GP129 chemokine mutant virus pathogenicity study demonstrated reduced viral load to target organs but highly extended viremia.
Keywords: Congenital infection; Cytomegalovirus; Epithelial cells; Glycoproteins; Guinea pig; Pentameric complex; Placenta; Trophoblast; UL128; Viral tropism.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures








Similar articles
-
A Homolog Pentameric Complex Dictates Viral Epithelial Tropism, Pathogenicity and Congenital Infection Rate in Guinea Pig Cytomegalovirus.PLoS Pathog. 2016 Jul 7;12(7):e1005755. doi: 10.1371/journal.ppat.1005755. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27387220 Free PMC article.
-
Repair of a Mutation Disrupting the Guinea Pig Cytomegalovirus Pentameric Complex Acquired during Fibroblast Passage Restores Pathogenesis in Immune-Suppressed Guinea Pigs and in the Context of Congenital Infection.J Virol. 2016 Aug 12;90(17):7715-27. doi: 10.1128/JVI.00320-16. Print 2016 Sep 1. J Virol. 2016. PMID: 27307567 Free PMC article.
-
Inclusion of the Guinea Pig Cytomegalovirus Pentameric Complex in a Live Virus Vaccine Aids Efficacy against Congenital Infection but Is Not Essential for Improving Maternal and Neonatal Outcomes.Viruses. 2021 Nov 26;13(12):2370. doi: 10.3390/v13122370. Viruses. 2021. PMID: 34960639 Free PMC article.
-
Characteristics and functions of human cytomegalovirus UL128 gene/protein.Acta Virol. 2014;58(2):103-7. doi: 10.4149/av_2014_02_103. Acta Virol. 2014. PMID: 24957713 Review.
-
Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications.Rev Med Virol. 2010 May;20(3):136-55. doi: 10.1002/rmv.645. Rev Med Virol. 2010. PMID: 20084641 Review.
Cited by
-
Guinea pig cytomegalovirus protective T cell antigen GP83 is a functional pp65 homolog for innate immune evasion and pentamer dependent virus tropism.J Virol. 2021 Apr 26;95(10):e00324-21. doi: 10.1128/JVI.00324-21. Epub 2021 Mar 3. J Virol. 2021. PMID: 33658350 Free PMC article.
-
Neutralizing antibodies to gB based CMV vaccine requires full length antigen but reduced virus neutralization on non-fibroblast cells limits vaccine efficacy in the guinea pig model.Vaccine. 2020 Feb 28;38(10):2340-2349. doi: 10.1016/j.vaccine.2020.01.063. Epub 2020 Jan 31. Vaccine. 2020. PMID: 32008881 Free PMC article.
-
Inclusion of the Viral Pentamer Complex in a Vaccine Design Greatly Improves Protection against Congenital Cytomegalovirus in the Guinea Pig Model.J Virol. 2019 Oct 29;93(22):e01442-19. doi: 10.1128/JVI.01442-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31484753 Free PMC article.
-
The pentameric complex is not required for congenital CMV transmission in seronegative rhesus macaques.Sci Transl Med. 2025 Mar 12;17(789):eadm8961. doi: 10.1126/scitranslmed.adm8961. Epub 2025 Mar 12. Sci Transl Med. 2025. PMID: 40073152 Free PMC article.
-
Endothelial Cell Infection by Guinea Pig Cytomegalovirus Is a Lytic or Persistent Infection Depending on Tissue Origin but Requires Viral Pentamer Complex and pp65 Tegument Protein.J Virol. 2022 Sep 14;96(17):e0083122. doi: 10.1128/jvi.00831-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000848 Free PMC article.
References
-
- Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. Genetic content of wild-type human cytomegalovirus. J Gen Virol. 2004;85(Pt 5):1301–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

