Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway
- PMID: 28651608
- PMCID: PMC5485714
- DOI: 10.1186/s12943-017-0677-9
Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway
Retraction in
-
Retraction Note to: Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway.Mol Cancer. 2022 Sep 29;21(1):188. doi: 10.1186/s12943-022-01660-3. Mol Cancer. 2022. PMID: 36175912 Free PMC article. No abstract available.
Abstract
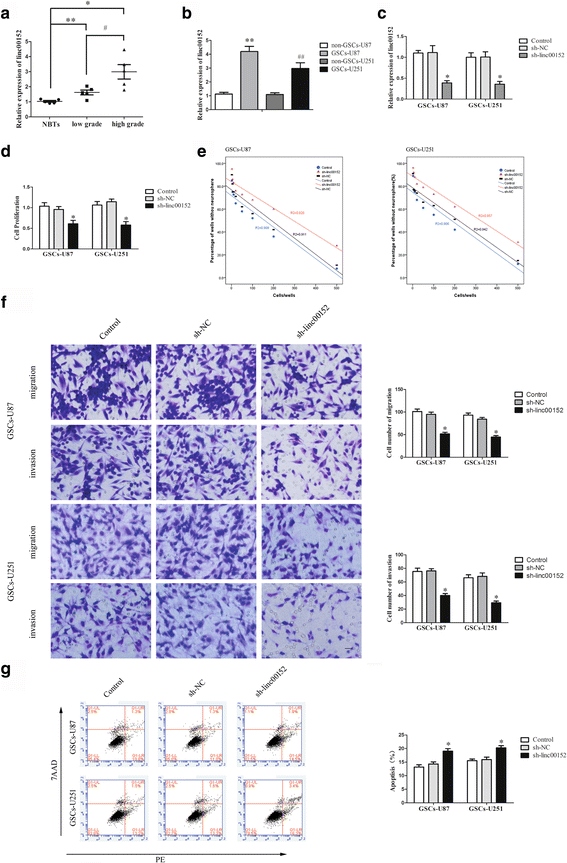
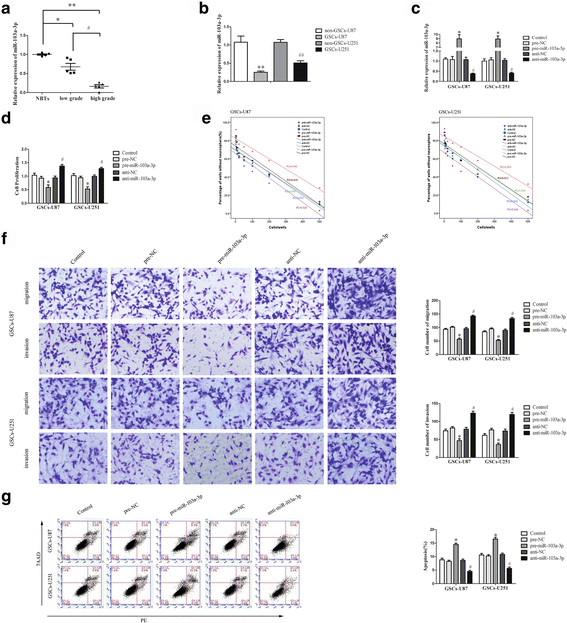
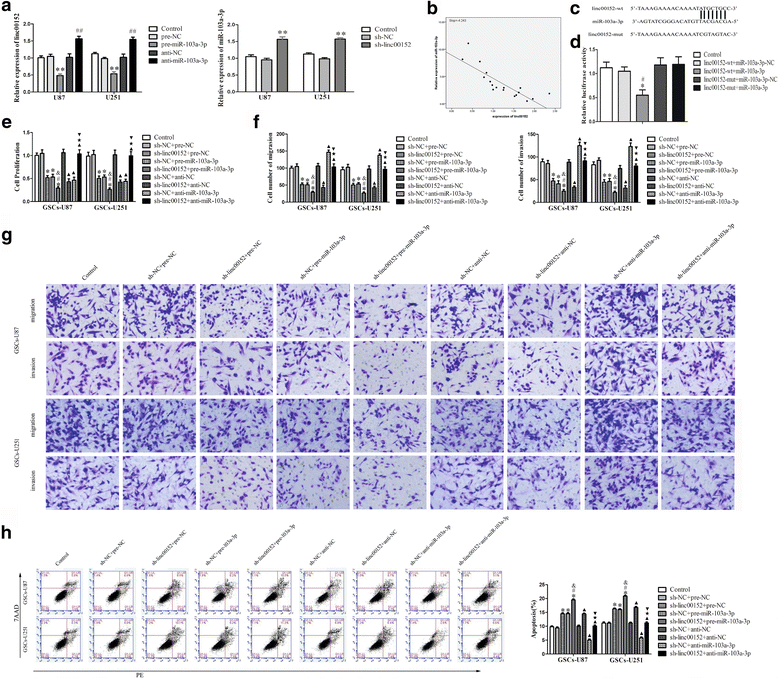
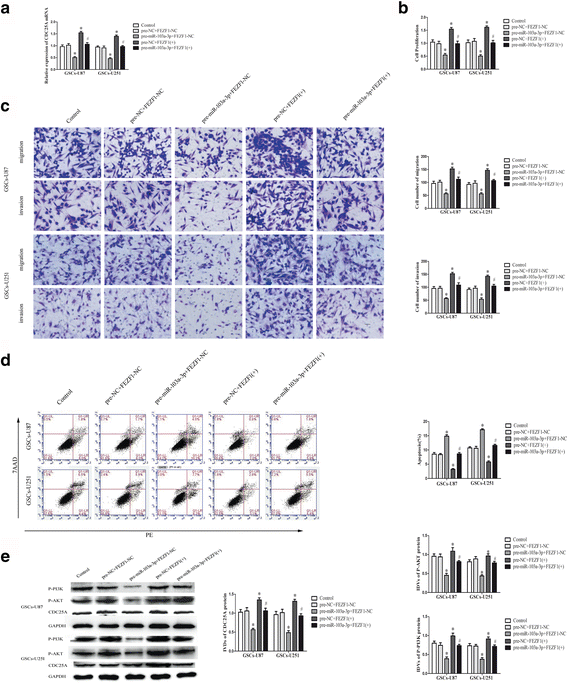
Background: Glioma is one of the most frequent intracranial malignant tumors. LncRNAs have been identified as new modulators in the origination and progression of glioma.
Methods: Quantitative real-time PCR were conducted to evaluate the expression of linc00152 and miRNA-103a-3p in glioma tissues and cells. Western blot were used to determine the expression of FEZF1 and CDC25A in glioma tissues and cells. Stable knockdown of linc00152 or over-expression of miR-103a-3p in glioma stem cells (GSCs) were established to explore the function of linc00152 and miR-103a-3p in GSCs. Further, luciferase reports were used to investigate the correlation between linc00152 and miR-103a-3p. Cell Counting Kit-8, transwell assays, and flow cytometry were used to investigate the function of linc00152 and miR-103a-3p in GSC malignant biological behaviors. ChIP assays were employed to ascertain the correlations between FEZF1 and CDC25A.
Results: Linc00152 was up-regulated in glioma tissues as well as in GSCs. Knockdown of linc00152 inhibited cell proliferation, migration and invasion, while promoted GSC apoptosis. Linc00152 regulated the malignant behavior of GSCs by binding to miR-103a-3p, which functions as a tumor suppressor. In addition, knockdown of linc00152 down-regulated forebrain embryonic zinc finger protein 1 (FEZF1), a direct target of miR-103a-3p which played an oncogenic role in GSCs. FEZF1 elevated promoter activities and up-regulated expression of the oncogenic gene cell division cycle 25A (CDC25A). CDC25A over-expression activated the PI3K/AKT pathways, which regulated the malignant behavior of GSCs.
Conclusions: Linc00152/miR-103a-3p/FEZF1/CDC25A axis plays a novel role in regulating the malignant behavior of GSCs, which may be a new potential therapeutic strategy for glioma therapy.
Keywords: CDC25A; FEZF1; Glioma stem cells; Long non-coding RNA; linc00152; miR-103a-3p.
Figures




Similar articles
-
Long non-coding RNA LINC00152/miR-613/CD164 axis regulates cell proliferation, apoptosis, migration and invasion in glioma via PI3K/AKT pathway.Neoplasma. 2020 Jul;67(4):762-772. doi: 10.4149/neo_2020_190706N598. Epub 2020 Jul 23. Neoplasma. 2020. PMID: 32726126
-
Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122.Mol Cancer. 2017 Nov 13;16(1):171. doi: 10.1186/s12943-017-0737-1. Mol Cancer. 2017. PMID: 29132362 Free PMC article.
-
MOV10 binding circ-DICER1 regulates the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4 expression change.J Exp Clin Cancer Res. 2019 Jan 8;38(1):9. doi: 10.1186/s13046-018-0990-1. J Exp Clin Cancer Res. 2019. PMID: 30621721 Free PMC article.
-
LINC00152 knockdown exerts repressive effects on epithelial-mesenchymal transition in bladder cancer.Cytotechnology. 2025 Oct;77(5):166. doi: 10.1007/s10616-025-00829-0. Epub 2025 Aug 20. Cytotechnology. 2025. PMID: 40851931
-
The role of microRNAs in glioma initiation and progression.Front Biosci (Landmark Ed). 2012 Jan 1;17(2):700-12. doi: 10.2741/3952. Front Biosci (Landmark Ed). 2012. PMID: 22201769 Free PMC article. Review.
Cited by
-
Long non-coding RNA LINC00152 in cancer: Roles, mechanisms, and chemotherapy and radiotherapy resistance.Front Oncol. 2022 Aug 10;12:960193. doi: 10.3389/fonc.2022.960193. eCollection 2022. Front Oncol. 2022. PMID: 36033524 Free PMC article. Review.
-
Non-coding RNAs and glioma: Focus on cancer stem cells.Mol Ther Oncolytics. 2022 Sep 17;27:100-123. doi: 10.1016/j.omto.2022.09.005. eCollection 2022 Dec 15. Mol Ther Oncolytics. 2022. PMID: 36321132 Free PMC article. Review.
-
Zebrafish xenograft model of human lung cancer for studying the function of LINC00152 in cell proliferation and invasion.Cancer Cell Int. 2020 Aug 6;20:376. doi: 10.1186/s12935-020-01460-z. eCollection 2020. Cancer Cell Int. 2020. PMID: 32774169 Free PMC article.
-
lncRNA CYTOR Facilitates Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Modulating SOX11 via Sponging miR-6512-3p.Stem Cells Int. 2023 Mar 3;2023:5671809. doi: 10.1155/2023/5671809. eCollection 2023. Stem Cells Int. 2023. PMID: 36910334 Free PMC article.
-
Targeting YAP1/LINC00152/FSCN1 Signaling Axis Prevents the Progression of Colorectal Cancer.Adv Sci (Weinh). 2019 Dec 5;7(3):1901380. doi: 10.1002/advs.201901380. eCollection 2020 Feb. Adv Sci (Weinh). 2019. PMID: 32042551 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

