Mammalian ECD Protein Is a Novel Negative Regulator of the PERK Arm of the Unfolded Protein Response
- PMID: 28652267
- PMCID: PMC5574048
- DOI: 10.1128/MCB.00030-17
Mammalian ECD Protein Is a Novel Negative Regulator of the PERK Arm of the Unfolded Protein Response
Abstract
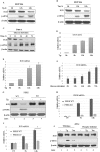
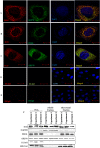
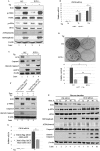
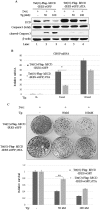
Mammalian Ecdysoneless (ECD) is a highly conserved ortholog of the DrosophilaEcd gene product whose mutations impair the synthesis of Ecdysone and produce cell-autonomous survival defects, but the mechanisms by which ECD functions are largely unknown. Here we present evidence that ECD regulates the endoplasmic reticulum (ER) stress response. ER stress induction led to a reduced ECD protein level, but this effect was not seen in PKR-like ER kinase knockout (PERK-KO) or phosphodeficient eukaryotic translation initiation factor 2α (eIF2α) mouse embryonic fibroblasts (MEFs); moreover, ECD mRNA levels were increased, suggesting impaired ECD translation as the mechanism for reduced protein levels. ECD colocalizes and coimmunoprecipitates with PERK and GRP78. ECD depletion increased the levels of both phospho-PERK (p-PERK) and p-eIF2α, and these effects were enhanced upon ER stress induction. Reciprocally, overexpression of ECD led to marked decreases in p-PERK, p-eIF2α, and ATF4 levels but robust increases in GRP78 protein levels. However, GRP78 mRNA levels were unchanged, suggesting a posttranscriptional event. Knockdown of GRP78 reversed the attenuating effect of ECD overexpression on PERK signaling. Significantly, overexpression of ECD provided a survival advantage to cells upon ER stress induction. Taken together, our data demonstrate that ECD promotes survival upon ER stress by increasing GRP78 protein levels to enhance the adaptive folding protein in the ER to attenuate PERK signaling.
Keywords: ECD; ER; GRP78; PERK; UPR; cell survival.
Copyright © 2017 Olou et al.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
