Immune Escape in Breast Cancer During In Situ to Invasive Carcinoma Transition
- PMID: 28652380
- PMCID: PMC5628128
- DOI: 10.1158/2159-8290.CD-17-0222
Immune Escape in Breast Cancer During In Situ to Invasive Carcinoma Transition
Abstract
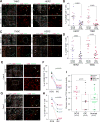
To investigate immune escape during breast tumor progression, we analyzed the composition of leukocytes in normal breast tissues, ductal carcinoma in situ (DCIS), and invasive ductal carcinomas (IDC). We found significant tissue and tumor subtype-specific differences in multiple cell types including T cells and neutrophils. Gene expression profiling of CD45+CD3+ T cells demonstrated a decrease in CD8+ signatures in IDCs. Immunofluorescence analysis showed fewer activated GZMB+CD8+ T cells in IDC than in DCIS, including in matched DCIS and recurrent IDC. T-cell receptor clonotype diversity was significantly higher in DCIS than in IDCs. Immune checkpoint protein TIGIT-expressing T cells were more frequent in DCIS, whereas high PD-L1 expression and amplification of CD274 (encoding PD-L1) was only detected in triple-negative IDCs. Coamplification of a 17q12 chemokine cluster with ERBB2 subdivided HER2+ breast tumors into immunologically and clinically distinct subtypes. Our results show coevolution of cancer cells and the immune microenvironment during tumor progression.Significance: The design of effective cancer immunotherapies requires the understanding of mechanisms underlying immune escape during tumor progression. Here we demonstrate a switch to a less active tumor immune environment during the in situ to invasive breast carcinoma transition, and identify immune regulators and genomic alterations that shape tumor evolution. Cancer Discov; 7(10); 1098-115. ©2017 AACR.See related commentary by Speiser and Verdeil, p. 1062This article is highlighted in the In This Issue feature, p. 1047.
©2017 American Association for Cancer Research.
Conflict of interest statement
Figures




Comment in
-
More T Cells versus Better T Cells in Patients with Breast Cancer.Cancer Discov. 2017 Oct;7(10):1062-1064. doi: 10.1158/2159-8290.CD-17-0858. Cancer Discov. 2017. PMID: 28974529
References
-
- Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–41. - PubMed
-
- de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. - PubMed
-
- Schedin P, O'Brien J, Rudolph M, Stein T, Borges V. Microenvironment of the involuting mammary gland mediates mammary cancer progression. J Mammary Gland Biol Neoplasia. 2007;12:71–82. - PubMed
-
- Man YG, Sang QX. The significance of focal myoepithelial cell layer disruptions in human breast tumor invasion: a paradigm shift from the “protease-centered” hypothesis. Exp Cell Res. 2004;301:103–18. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

