Replication Study: The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate
- PMID: 28653623
- PMCID: PMC5487214
- DOI: 10.7554/eLife.26030
Replication Study: The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate
Abstract
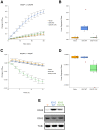
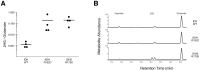
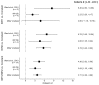
In 2016, as part of the Reproducibility Project: Cancer Biology, we published a Registered Report (Fiehn et al., 2016), that described how we intended to replicate selected experiments from the paper "The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate" (Ward et al., 2010). Here, we report the results of those experiments. We found that cells expressing R172K mutant IDH2 did not display isocitrate-dependent NADPH production above vector control levels, in contrast to the increased production observed with wild-type IDH2. Conversely, expression of R172K mutant IDH2 resulted in increased alpha-ketoglutarate-dependent consumption of NADPH compared to wild-type IDH2 or vector control. These results are similar to those reported in the original study (Figure 2; Ward et al., 2010). Further, expression of R172K mutant IDH2 resulted in increased 2HG levels within cells compared to the background levels observed in wild-type IDH2 and vector control, similar to the original study (Figure 3D; Ward et al., 2010). In primary human AML samples, the 2HG levels observed in samples with mutant IDH1 or IDH2 status were higher than those observed in samples without an IDH mutation, similar to what was observed in the original study (Figure 5C; Ward et al., 2010). Finally, we report meta-analyses for each result.
Keywords: 2-HG; Reproducibility Project: Cancer Biology; acute myeloid leukemia; biochemistry; cancer biology; human; metascience; replication; reproducibility.
Conflict of interest statement
MRS: West Coast Metabolomics Center, University of California, Davis is a Science Exchange associated lab.
TC: West Coast Metabolomics Center, University of California, Davis is a Science Exchange associated lab.
OF: West Coast Metabolomics Center, University of California, Davis is a Science Exchange associated lab.
RP:CB: EI, NP: Employed by and hold shares in Science Exchange Inc.
The other authors declare that no competing interests exist.
Figures



References
-
- Allwood JW, Erban A, de Koning S, Dunn WB, Luedemann A, Lommen A, Kay L, Löscher R, Kopka J, Goodacre R. Inter-laboratory reproducibility of fast gas chromatography-electron impact-time of flight mass spectrometry (GC-EI-TOF/MS) based plant metabolomics. Metabolomics. 2009;5:479–496. doi: 10.1007/s11306-009-0169-z. - DOI - PMC - PubMed
-
- Borger DR, Goyal L, Yau T, Poon RT, Ancukiewicz M, Deshpande V, Christiani DC, Liebman HM, Yang H, Kim H, Yen K, Faris JE, Iafrate AJ, Kwak EL, Clark JW, Allen JN, Blaszkowsky LS, Murphy JE, Saha SK, Hong TS, Wo JY, Ferrone CR, Tanabe KK, Bardeesy N, Straley KS, Agresta S, Schenkein DP, Ellisen LW, Ryan DP, Zhu AX. Circulating oncometabolite 2-hydroxyglutarate is a potential surrogate biomarker in patients with isocitrate dehydrogenase-mutant intrahepatic cholangiocarcinoma. Clinical Cancer Research. 2014;20:1884–1890. doi: 10.1158/1078-0432.CCR-13-2649. - DOI - PMC - PubMed
-
- Carbonneau M, M Gagné L, Lalonde ME, Germain MA, Motorina A, Guiot MC, Secco B, Vincent EE, Tumber A, Hulea L, Bergeman J, Oppermann U, Jones RG, Laplante M, Topisirovic I, Petrecca K, Huot MÉ, Mallette FA, Gagné M, Guiot A, Secco M-C, Tumber EE, Jones U, Huot K. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nature Communications. 2016;7:12700. doi: 10.1038/ncomms12700. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

