Comprehensive DNA Methylation Analysis Using a Methyl-CpG-binding Domain Capture-based Method in Chronic Lymphocytic Leukemia Patients
- PMID: 28654059
- PMCID: PMC5608441
- DOI: 10.3791/55773
Comprehensive DNA Methylation Analysis Using a Methyl-CpG-binding Domain Capture-based Method in Chronic Lymphocytic Leukemia Patients
Abstract
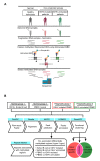
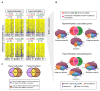
The role of long noncoding RNAs (lncRNAs) in cancer is coming to the forefront due to growing interest in understanding their mechanistic functions during cancer development and progression. Despite this, the global epigenetic regulation of lncRNAs and repetitive sequences in cancer has not been well investigated, particularly in chronic lymphocytic leukemia (CLL). This study focuses on a unique approach: the immunoprecipitation-based capture of double-stranded, methylated DNA fragments using methyl-binding domain (MBD) proteins, followed by next-generation sequencing (MBD-seq). CLL patient samples belonging to two prognostic subgroups (5 IGVH mutated samples + 5 IGVH unmutated samples) were used in this study. Analysis revealed 5,800 hypermethylated and 12,570 hypomethylated CLL-specific differentially methylated genes (cllDMGs) compared to normal healthy controls. Importantly, these results identified several CLL-specific, differentially methylated lncRNAs, repetitive elements, and protein-coding genes with potential prognostic value. This work outlines a detailed protocol for an MBD-seq and bioinformatics pipeline developed for the comprehensive analysis of global methylation profiles in highly CpG-rich regions using CLL patient samples. Finally, a protein-coding gene and an lncRNA were validated using pyrosequencing, which is a highly quantitative method to analyze CpG methylation levels to further corroborate the findings from the MBD-seq protocol.
References
-
- Kanduri M, et al. Differential genome-wide array-based methylation profiles in prognostic subsets of chronic lymphocytic leukemia. Blood. 2010;115(2):296–305. - PubMed
-
- Cahill N, et al. 450K-array analysis of chronic lymphocytic leukemia cells reveals global DNA methylation to be relatively stable over time and similar in resting and proliferative compartments. Leukemia. 2013;27(1):150–158. - PubMed
-
- Kulis M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236–1242. - PubMed
-
- Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336(6083):934–937. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources