Transcriptome-wide piRNA profiling in human brains of Alzheimer's disease
- PMID: 28654860
- PMCID: PMC5542056
- DOI: 10.1016/j.neurobiolaging.2017.05.020
Transcriptome-wide piRNA profiling in human brains of Alzheimer's disease
Abstract
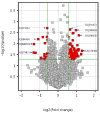
Discovered in the brains of multiple animal species, piRNAs may contribute to the pathogenesis of neuropsychiatric illnesses. The present study aimed to identify brain piRNAs across transcriptome that are associated with Alzheimer's disease (AD). Prefrontal cortical tissues of 6 AD cases and 6 controls were examined for piRNA expression levels using an Arraystar HG19 piRNA array (containing 23,677 piRNAs) and genotyped for 17 genome-wide significant and replicated risk SNPs. We examined whether piRNAs are expressed differently between AD cases and controls and explored the potential regulatory effects of risk SNPs on piRNA expression levels. We identified a total of 9453 piRNAs in human brains, with 103 nominally (p < 0.05) differentially (>1.5 fold) expressed in AD cases versus controls and most of the 103 piRNAs nominally correlated with genome-wide significant risk SNPs. We conclude that piRNAs are abundant in human brains and may represent risk biomarkers of AD.
Keywords: Alzheimer's disease; Brain; Differential expression; Microarray; Transcriptome; piRNA.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
References
-
- Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O’Donovan MC, Williams J, Owen MJ, Kirov G. A genome-wide association study for late-onset Alzheimer’s disease using DNA pooling. BMC Med Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. - DOI - PMC - PubMed
-
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129(4):965–72. - PubMed
-
- Antunez C, Boada M, Gonzalez-Perez A, Gayan J, Ramirez-Lorca R, Marin J, Hernandez I, Moreno-Rey C, Moron FJ, Lopez-Arrieta J, Mauleon A, Rosende-Roca M, Noguera-Perea F, Legaz-Garcia A, Vivancos-Moreau L, Velasco J, Carrasco JM, Alegret M, Antequera-Torres M, Manzanares S, Romo A, Blanca I, Ruiz S, Espinosa A, Castano S, Garcia B, Martinez-Herrada B, Vinyes G, Lafuente A, Becker JT, Galan JJ, Serrano-Rios M, Vazquez E, Tarraga L, Saez ME, Lopez OL, Real LM, Ruiz A Alzheimer’s Disease Neuroimaging I. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer’s disease. Genome Med. 2011;3(5):33. doi: 10.1186/gm249. - DOI - PMC - PubMed
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–7. doi: 10.1038/nature04916. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

