Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens
- PMID: 28655737
- PMCID: PMC5555476
- DOI: 10.1534/g3.117.041277
Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens
Abstract
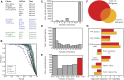
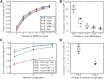
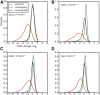
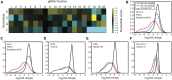
The adaptation of CRISPR/SpCas9 technology to mammalian cell lines is transforming the study of human functional genomics. Pooled libraries of CRISPR guide RNAs (gRNAs) targeting human protein-coding genes and encoded in viral vectors have been used to systematically create gene knockouts in a variety of human cancer and immortalized cell lines, in an effort to identify whether these knockouts cause cellular fitness defects. Previous work has shown that CRISPR screens are more sensitive and specific than pooled-library shRNA screens in similar assays, but currently there exists significant variability across CRISPR library designs and experimental protocols. In this study, we reanalyze 17 genome-scale knockout screens in human cell lines from three research groups, using three different genome-scale gRNA libraries. Using the Bayesian Analysis of Gene Essentiality algorithm to identify essential genes, we refine and expand our previously defined set of human core essential genes from 360 to 684 genes. We use this expanded set of reference core essential genes, CEG2, plus empirical data from six CRISPR knockout screens to guide the design of a sequence-optimized gRNA library, the Toronto KnockOut version 3.0 (TKOv3) library. We then demonstrate the high effectiveness of the library relative to reference sets of essential and nonessential genes, as well as other screens using similar approaches. The optimized TKOv3 library, combined with the CEG2 reference set, provide an efficient, highly optimized platform for performing and assessing gene knockout screens in human cell lines.
Keywords: CRISPR/Cas9; cancer cell lines; core essential genes; genetic screens.
Copyright © 2017 Hart et al.
Figures
References
-
- Blomen V. A., Majek P., Jae L. T., Bigenzahn J. W., Nieuwenhuis J., et al. , 2015. Gene essentiality and synthetic lethality in haploid human cells. Science 350: 1092–1096. - PubMed
-
- Echeverri C. J., Beachy P. A., Baum B., Boutros M., Buchholz F., et al. , 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods 3: 777–779. - PubMed
-
- Evers B., Jastrzebski K., Heijmans J. P., Grernrum W., Beijersbergen R. L., et al. , 2016. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 34: 631–633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
