Human Cytomegalovirus-Infected Glioblastoma Cells Display Stem Cell-Like Phenotypes
- PMID: 28656174
- PMCID: PMC5480031
- DOI: 10.1128/mSphere.00137-17
Human Cytomegalovirus-Infected Glioblastoma Cells Display Stem Cell-Like Phenotypes
Abstract
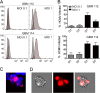
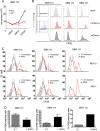
Glioblastoma multiforme (GBM) is the most common brain tumor in adults. Human cytomegalovirus (HCMV) genomes are present in GBM tumors, yielding hope that antiviral treatments could prove therapeutic and improve the poor prognosis of GBM patients. We discovered that GBM cells infected in vitro with HCMV display properties of cancer stem cells. HCMV-infected GBM cells grow more slowly than mock-infected controls, demonstrate a higher capacity for self-renewal determined by a sphere formation assay, and display resistance to the chemotherapeutic drug temozolomide. Our data suggest that HCMV, while present in only a minority of the cells within a tumor, could contribute to the pathogenesis of GBMs by promoting or prolonging stem cell-like phenotypes, thereby perpetuating tumors in the face of chemotherapy. Importantly, we show that temozolomide sensitivity is restored by the antiviral drug ganciclovir, indicating a potential mechanism underlying the positive effects observed in GBM patients treated with antiviral therapy. IMPORTANCE A role for HCMV in GBMs remains controversial for several reasons. Some studies find HCMV in GBM tumors, while others do not. Few cells within a GBM may harbor HCMV, making it unclear how the virus could be contributing to the tumor phenotype without infecting every cell. Finally, HCMV does not overtly transform cells in vitro. However, tumors induced by other viruses can be treated with antiviral remedies, and initial results indicate that this may be true for anti-HCMV therapies and GBMs. With such a poor prognosis for GBM patients, any potential new intervention deserves exploration. Our work here describes an evidence-based model for how HCMV could contribute to GBM biology while infecting very few cells and without transforming them. It also illuminates why anti-HCMV treatments may be beneficial to GBM patients. Our observations provide blueprints for future in vitro studies examining how HCMV manipulates stem cell-specific pathways and future clinical studies of anti-HCMV measures as GBM therapeutics.
Keywords: brain cancer; cancer; chemoresistance; chemotherapy; herpesvirus.
Figures





References
-
- Hariri S, Bennett NM, Niccolai LM, Schafer S, Park IU, Bloch KC, Unger ER, Whitney E, Julian P, Scahill MW, Abdullah N, Levine D, Johnson ML, Steinau M, Markowitz LE, HPV-IMPACT Working Group . 2015. Reduction in HPV 16/18-associated high grade cervical lesions following HPV vaccine introduction in the United States − 2008 2012. Vaccine 33:1608–1613. doi: 10.1016/j.vaccine.2015.01.084. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

