Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics
- PMID: 28656958
- PMCID: PMC5493757
- DOI: 10.1038/ncomms16054
Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics
Abstract
Establishing virus-host relationships has historically relied on culture-dependent approaches. Here we report on the use of marine metatranscriptomics to probe virus-host relationships. Statistical co-occurrence analyses of dsDNA, ssRNA and dsRNA viral markers of polyadenylation-selected RNA sequences from microbial communities dominated by Aureococcus anophagefferens (Quantuck Bay, NY), and diatoms (Narragansett Bay, RI) show active infections by diverse giant viruses (NCLDVs) associated with algal and nonalgal hosts. Ongoing infections of A. anophagefferens by a known Mimiviridae (AaV) occur during bloom peak and decline. Bloom decline is also accompanied by increased activity of viruses other than AaV, including (+) ssRNA viruses. In Narragansett Bay, increased temporal resolution reveals active NCLDVs with both 'boom-and-bust' and 'steady-state infection'-like ecologies that include known as well as novel virus-host interactions. Our approach offers a method for screening active viral infections and develops links between viruses and their potential hosts in situ. Our observations further demonstrate that previously unknown virus-host relationships in marine systems are abundant.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

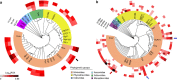
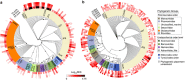


References
-
- Breitbart M. Marine viruses: truth or dare. Ann. Rev. Mar. Sci. 4, 425–448 (2012). - PubMed
-
- Brum J. R. et al. Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015). - PubMed
-
- Short S. M. The ecology of viruses that infect eukaryotic algae. Environ. Microbiol. 14, 2253–2271 (2012). - PubMed
-
- Ogata H., Monier A. & Claverie J.-M. in Global Change: Mankind-Marine Environment Interactions eds Ceccaldi H., Dekeyser I., Girault M., Stora G. 157–162Springer Netherlands (2011).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

