Cooperative RNA Folding under Cellular Conditions Arises From Both Tertiary Structure Stabilization and Secondary Structure Destabilization
- PMID: 28657303
- PMCID: PMC5542450
- DOI: 10.1021/acs.biochem.7b00325
Cooperative RNA Folding under Cellular Conditions Arises From Both Tertiary Structure Stabilization and Secondary Structure Destabilization
Abstract
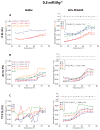
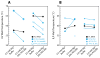
RNA folding has been studied extensively in vitro, typically under dilute solution conditions and abiologically high salt concentrations of 1 M Na+ or 10 mM Mg2+. The cellular environment is very different, with 20-40% crowding and only 10-40 mM Na+, 140 mM K+, and 0.5-2.0 mM Mg2+. As such, RNA structures and functions can be radically altered under cellular conditions. We previously reported that tRNAphe secondary and tertiary structures unfold together in a cooperative two-state fashion under crowded in vivo-like ionic conditions, but in a noncooperative multistate fashion under dilute in vitro ionic conditions unless in nonphysiologically high concentrations of Mg2+. The mechanistic basis behind these effects remains unclear, however. To address the mechanism that drives RNA folding cooperativity, we probe effects of cellular conditions on structures and stabilities of individual secondary structure fragments comprising the full-length RNA. We elucidate effects of a diverse set of crowders on tRNA secondary structural fragments and full-length tRNA at three levels: at the nucleotide level by temperature-dependent in-line probing, at the tertiary structure level by small-angle X-ray scattering, and at the global level by thermal denaturation. We conclude that cooperative RNA folding is induced by two overlapping mechanisms: increased stability and compaction of tertiary structure through effects of Mg2+, and decreased stability of certain secondary structure elements through the effects of molecular crowders. These findings reveal that despite having very different chemical makeups RNA and protein can both have weak secondary structures in vivo leading to cooperative folding.
Figures






Similar articles
-
Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions.RNA. 2014 Mar;20(3):331-47. doi: 10.1261/rna.042747.113. Epub 2014 Jan 17. RNA. 2014. PMID: 24442612 Free PMC article.
-
Altering the intermediate in the equilibrium folding of unmodified yeast tRNAPhe with monovalent and divalent cations.Biochemistry. 2001 Mar 27;40(12):3629-38. doi: 10.1021/bi002646+. Biochemistry. 2001. PMID: 11297430
-
Molecular Mechanism for Folding Cooperativity of Functional RNAs in Living Organisms.Biochemistry. 2018 May 22;57(20):2994-3002. doi: 10.1021/acs.biochem.8b00345. Epub 2018 May 7. Biochemistry. 2018. PMID: 29733204 Free PMC article.
-
Ribosomal binding of modified tRNA anticodons related to thermal stability.Nucleic Acids Symp Ser. 1997;(36):58-60. Nucleic Acids Symp Ser. 1997. PMID: 9478206 Review.
-
Circularly permuted DNA, RNA and proteins--a review.Gene. 1993 Mar 30;125(2):111-4. doi: 10.1016/0378-1119(93)90317-v. Gene. 1993. PMID: 7681803 Review.
Cited by
-
Single-nucleotide control of tRNA folding cooperativity under near-cellular conditions.Proc Natl Acad Sci U S A. 2019 Nov 12;116(46):23075-23082. doi: 10.1073/pnas.1913418116. Epub 2019 Oct 30. Proc Natl Acad Sci U S A. 2019. PMID: 31666318 Free PMC article.
-
Challenges and approaches to predicting RNA with multiple functional structures.RNA. 2018 Dec;24(12):1615-1624. doi: 10.1261/rna.067827.118. Epub 2018 Aug 24. RNA. 2018. PMID: 30143552 Free PMC article. Review.
-
In silico discovery of compounds targeting NPSL2: a regulatory element in the human oncomiR-1 primary microRNA.bioRxiv [Preprint]. 2025 May 9:2025.05.07.652683. doi: 10.1101/2025.05.07.652683. bioRxiv. 2025. PMID: 40654985 Free PMC article. Preprint.
-
In vivo-like nearest neighbor parameters improve prediction of fractional RNA base-pairing in cells.Nucleic Acids Res. 2023 Nov 10;51(20):11298-11317. doi: 10.1093/nar/gkad807. Nucleic Acids Res. 2023. PMID: 37855684 Free PMC article.
-
Differences in ion-RNA binding modes due to charge density variations explain the stability of RNA in monovalent salts.Sci Adv. 2022 Jul 22;8(29):eabo1190. doi: 10.1126/sciadv.abo1190. Epub 2022 Jul 20. Sci Adv. 2022. PMID: 35857829 Free PMC article.
References
-
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. - PubMed
-
- Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. - PubMed
-
- Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophil ribozyme. 1 Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

