16S sequencing and functional analysis of the fecal microbiome during treatment of newly diagnosed pediatric inflammatory bowel disease
- PMID: 28658154
- PMCID: PMC5500076
- DOI: 10.1097/MD.0000000000007347
16S sequencing and functional analysis of the fecal microbiome during treatment of newly diagnosed pediatric inflammatory bowel disease
Abstract
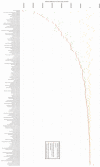
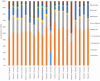
The human microbiome is of considerable interest to pediatric inflammatory bowel disease (PIBD) researchers with 1 potential mechanism for disease development being aberrant immune handling of the intestinal bacteria. This study analyses the fecal microbiome through treatment in newly diagnosed PIBD patients and compares to cohabiting siblings where possible. Patients were recruited on clinical suspicion of PIBD before diagnosis. Treatment-naïve fecal samples were collected, with further samples at 2 and 6 weeks into treatment. Samples underwent 16S ribosomal ribonucleic acid (RNA) gene sequencing and short-chain fatty acids (SCFAs) analysis, results were analyzed using quantitative-insights-into-microbial-ecology. Six PIBD patients were included in the cohort: 4 Crohn disease (CD), 1 ulcerative colitis (UC), 1 inflammatory bowel disease (IBD) unclassified, and median age 12.6 (range 10-15.1 years); 3 patients had an unaffected healthy sibling recruited. Microbial diversity (observed species/Chao1/Shannon diversity) was reduced in treatment-naïve patients compared to siblings and patients in remission. Principal coordinate analysis using Bray-Curtis dissimilarity and UniFrac revealed microbial shifts in CD over the treatment course. In treatment-naïve PIBD, there was reduction in functional ability for amino acid metabolism and carbohydrate handling compared to controls (P = .038) and patients in remission (P = .027). Metabolic function returned to normal after remission was achieved. SCFA revealed consistent detection of lactate in treatment-naïve samples. This study adds in-depth 16S rRNA sequencing analysis on a small longitudinal cohort to the literature and includes sibling controls and patients with UC/IBD unclassified. It highlights the initial dysbiosis, reduced diversity, altered functional potential, and subsequent shifts in bacteria from diagnosis over time to remission.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures
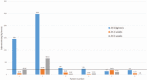
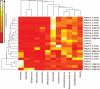
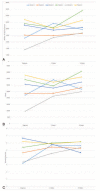
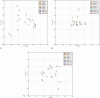
References
-
- Henderson P, van Limbergen JE, Wilson DC, et al. Genetics of childhood-onset inflammatory bowel disease. Inflamm Bowel Dis 2011;17:346–61. - PubMed
-
- Ashton JJ, Wiskin AE, Ennis S, et al. Rising incidence of paediatric inflammatory bowel disease (PIBD) in Wessex, Southern England. Arch Dis Child 2014;99:659–64. - PubMed
-
- Henderson P, Hansen R, Cameron FL, et al. Rising incidence of pediatric inflammatory bowel disease in Scotland. Inflamm Bowel Dis 2012;18:999–1005. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

